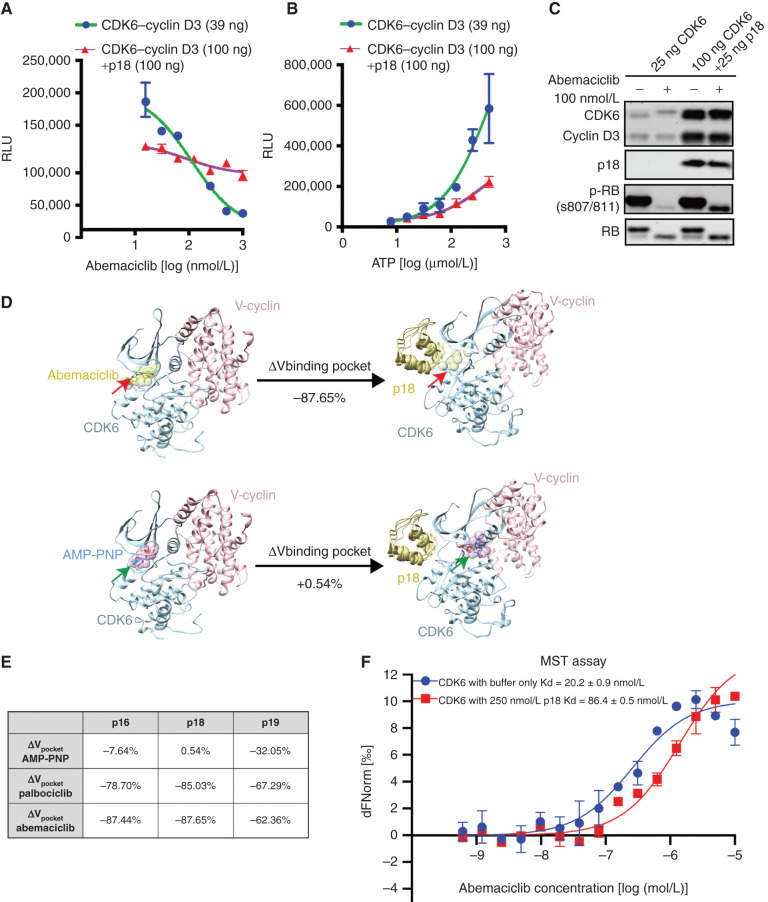
Figure 2.
INK4–CDK6 complexes are insensitive to CDK4/6i. A,In vitro kinase assay using recombinant CDK6–cyclin D3 and RB substrate demonstrates that preincubation of the complex with p18 (purple) prevents complete inhibition of kinase activity by abemaciclib. Data are shown as mean ± SD of two biological replicates. RLU, relative luminescence units. B, Effect of preincubation of p18 on CDK6–cyclin D3 in vitro kinase activity. Data are shown as mean ± SD of two biological replicates. C, Assay of CDK6–cyclin D3 kinase activity and response to p18 by immunoblotting demonstrating that p18 impairs the ability of abemaciclib to inhibit CDK6 phosphorylation of RB. D, Computational modeling of the effect of p18 binding to the CDK6 binding pocket expressed as volume change for abemaciclib (top) or AMP-PNP (bottom). Structures of CDK6–cyclin complex before and after p18 binding are represented by crystallographic structures with PDB IDs 2EUF and 1G3N (shown in ribbons). The binding pockets were approximated by spheres (shown in green and indicated by red arrows; shown in purple and indicated by green arrows). The volume of each binding pocket was quantified using the total volume of the corresponding set of spheres, and percentage of changes was calculated. E, The table summarizes the changes of binding pocket volume for two CDK4/6i (palbociclib and abemaciclib) and AMP-PNP upon binding of INK4s (p16, p18, p19). F, MST assay of CDK6 binding to abemaciclib showing the change in Kd as a result of p18 binding (red). Data are shown as mean ± SD of two independent measurements. See also Supplementary Fig. S2.