Figure 2. Discovery Approaches to Overcome Physiological Challenges and Control Microenvironment Interactions.
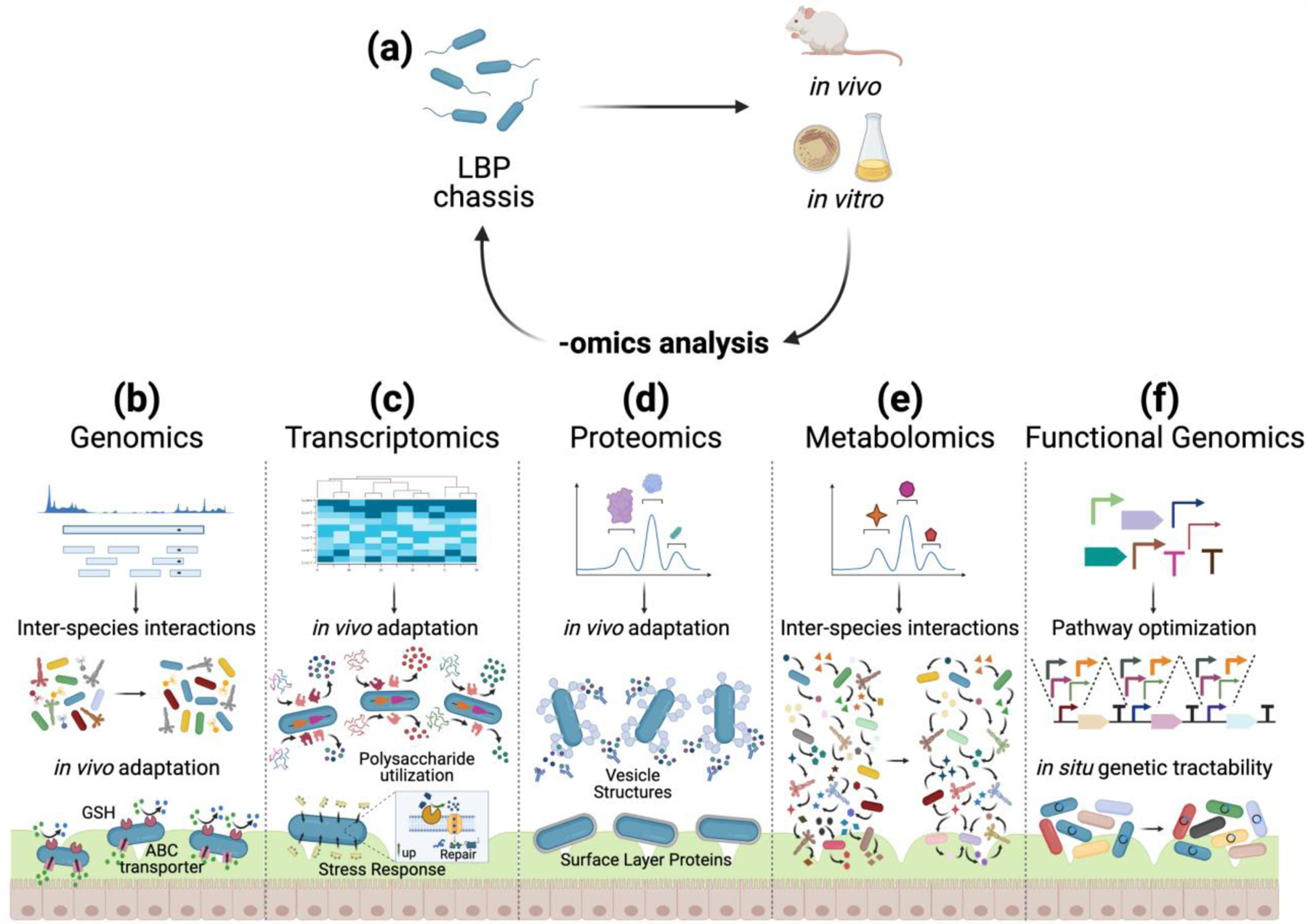
(a) Discovery of LBPs can be guided by investigating chassis in in vitro and in vivo settings, using a variety of ‘omics pipelines. (b) Genomics measures DNA composition at both the organism and community levels and can be used to determine the presence and abundance of LBPs in various locations in the gut and under various microenvironment conditions. These insights identify factors that drive the LBP’s adaptation to the gut and control interactions with other microbiota members. (c) Transcriptomics can identify genes that are differentially regulated in response to physiological challenges in the gut. These differentially regulated genes illuminate in vivo adaptation mechanisms of LBPs. (d) Proteomics can identify proteins in LBPs (as well as their subcellular localization) that facilitate in vivo adaptation. Since proteins on LBP surfaces mediate extracellular interactions, proteomics can provide insight into how LBPs interact and communicate with the dynamic gut microenvironment. (e) Metabolomics can identify the metabolic activity of LBPs in a multi-species community through identification of metabolites and their fluxes, which in turn illuminates how an LBP chassis adapts to gut microenvironments. (f) Functional genomics can be used to achieve pathway optimization and high-throughput strain screening for engineered LBP applications. Pathway optimization enables high activity of the engineered function even under the burden of physiological challenges. High-throughput strain screening can identify candidate chassis that are already gut colonizers and that can receive genetic payloads in situ through horizontal gene transfer.
