FIGURE 5: Neutral forces give rise to population bottlenecks during pore colonization.
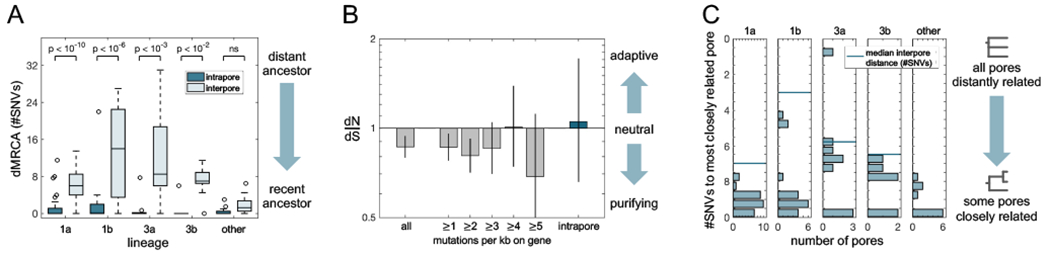
(A) The pattern of small intrapore dMRCAs (distance to the MRCA, averaged across colonies) as compared with interpore dMRCAs (average distance from the two pore MRCAs to their interpore MRCA) is consistent across subjects and lineages (Wilcoxon rank-sum test with Bonferroni correction). This indicates that pore populations are subject to recent strong bottlenecks and are ecologically isolated from each other. Analysis for (A) and (C) included all single pore samples (excluding hypermutators in Lineage 1a); pores from lineages containing fewer than five such samples are grouped as “other”. (B) Across all within-lineage SNVs, dN/dS (the ratio of nonsynonymous to synonymous mutations, relative to a neutral model; Methods) is slightly negative, indicative of purifying selection (P<0.0003). Values of dN/dS for genes with high mutational densities among within-lineage mutations are consistent with a neutral model, as is dN/dS for mutations inferred to have occurred inside pores. These findings suggest that non-adaptive evolution dominates C. acnes evolution on individuals and that adaptive sweeps are not responsible for low within-pore diversity. Error bars indicate 95% CIs. See Figure S12 for a more detailed analysis. (C) Pairs of pores on a person often share very recent common ancestry, suggesting that neutral bottlenecking occurred during a recent pore colonization or re-colonization event. The genetic distance between two pores is equal to twice the interpore dMRCA. Given that the number of pores sampled per subject was vastly smaller than the total number of pores on a person, these values underestimate the commonality of shared mutations between pore populations.
