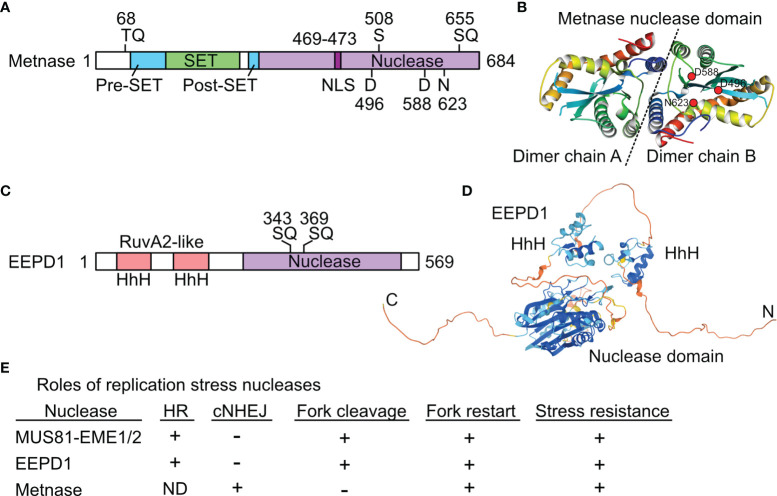
Figure 2.
Structures and roles of replication stress nucleases. (A) Metnase is a fusion of SET and nuclease domains. Two S/TQ sites (potential PIKK targets) are indicated, along with the DDN nuclease motif. S508 is phosphorylated by Chk1. (B) Crystal structure of Metnase nuclease domains shown as a dimer (separated by dashed line), as solved by the Georgiadis lab (59); image from the Protein Data Bank Japan (60) using the Molmil molecular structure viewer (61). Positions of DDN core nuclease residues are indicated in dimer chain B by red dots. (C) EEPD1 has two helix-hairpin-helix (HhH) domains related to prokaryotic RuvA2, a component of RuvAB that mediates Holliday junction branch migration. Two potential PIKK target SQ sites are indicated. (D) Predicted EEPD1 structure showing HhH and nuclease domains with intervening non-structured regions; image from AlphaFold (62). (E) Summary of known functions of three replication stress nucleases. ND, not determined; +, promotes process; -, not involved in indicated process. See text for further details.