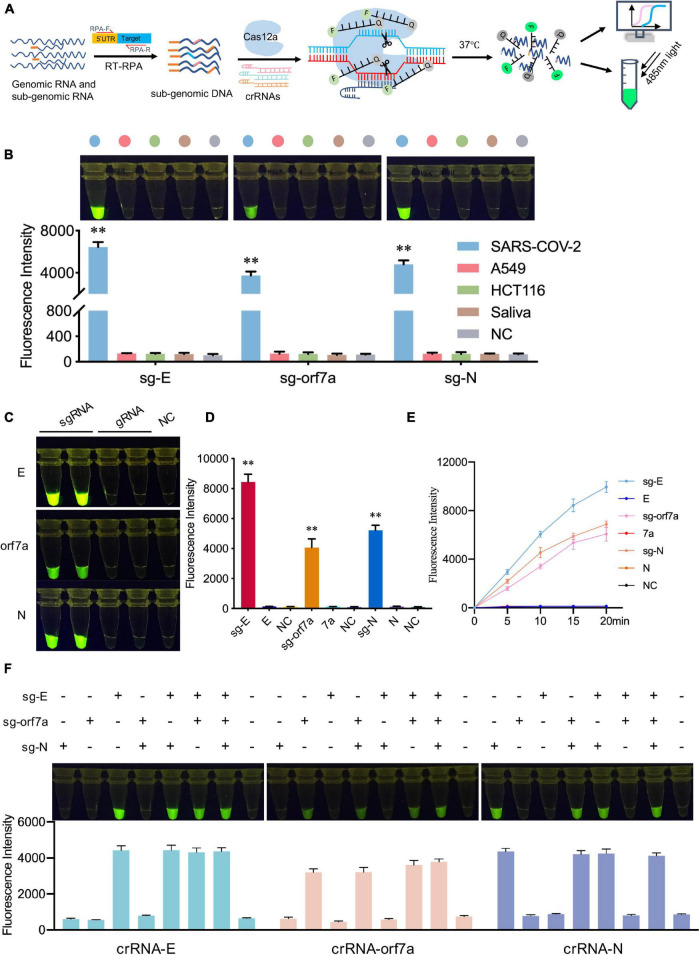
FIGURE 4.
CRISPR-actCoV can detect SARS-CoV-2 sgRNAs with high specificity. (A) Schematic illustration of the specificity assessment workflow. SARS-CoV-2 sgRNAs were amplified by RT-RPA using specific primers and subjected to CRISPR/Cas12a reporter system applied with specific crRNAs and ssDNA reporter. (B) Specificity validation of CRISPR-actCoV by distinguishing SARS-CoV-2 sgRNAs with background nucleic acid from human cell lines (A549, HCT116) or normal human saliva. The fluorescent images and intensities at 15 min of the reaction were shown. (C) Specificity assessment of CRISPR-actCoV by detecting samples containing SARS-CoV-2 sgRNA or gRNA. The fluorescent images at 15 min of the reaction were shown. (D) The fluorescent intensities of data in panel (C) were quantified by ImageJ and visualized with GraphPad. All the error bars were determined from three independent experiments. (E) A time-course assay of SARS-CoV-2 sgRNAs and gRNAs detection. (F) CRISPR-actCoV specificity assessment by detecting cross-reaction between different SARS-CoV-2 sgRNAs. sgRNA of E, orf7a, or N were detected individually or simultaneously in a reaction. The fluorescent images and intensities at 15 min of the reaction were shown. Data were collected from at least three independent experiments and was presented as Mean ± SD. The significance was considered as **p < 0.01.