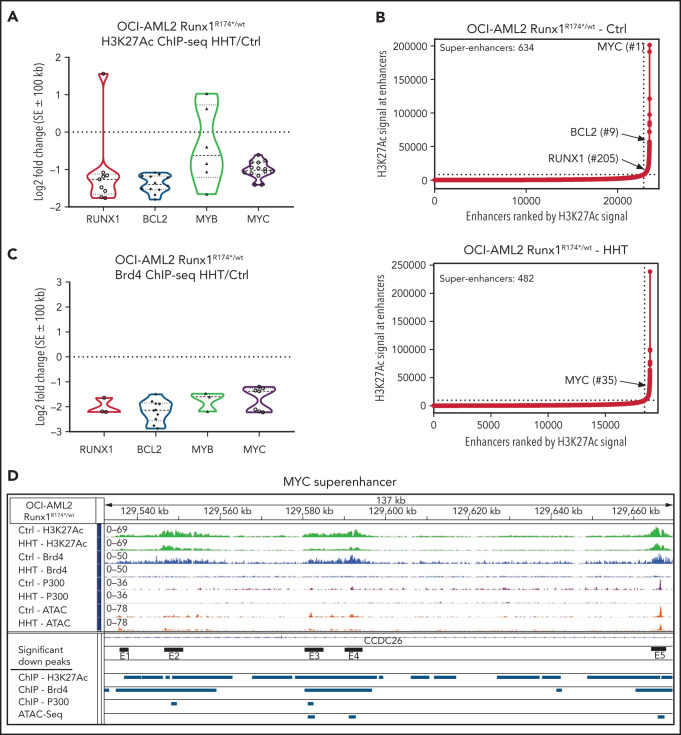
Figure 4.
Treatment with HHT alters the epigenome in mtRUNX1-expressing AML cells. (A) Log2 fold-change in H3K27Ac peaks (determined by ChIP-Seq analysis) at super enhancer +/− 100 kb in HHT-treated vs control OCI-AML2 RUNX1R174*/wt cells. Fold changes were calculated utilizing DiffReps. (B) OCI-AML2 RUNX1R174*/wt cells were treated with 100 nM of HHT for 16 hours and cross-linked for H3K27Ac ChIP analysis. Ranked ordering of super enhancers determined by H3K27Ac ChIP-Seq and ROSE analysis. (C) Log2 fold-change in BRD4 peaks (determined by ChIP-Seq analysis) at super enhancer +/− 100 kb in HHT-treated vs control OCI-AML2 RUNX1R174*/wt cells. Fold changes were calculated utilizing DiffReps. (D) IGV plot of sequence tag densities for H3K27Ac, BRD4 ChIP-Seq, P300 ChIP-Seq, and ATAC-Seq peaks at the MYC super-enhancer locus in HHT-treated vs control OCI-AML2 RUNX1R174*/wt cells. Fold changes were calculated utilizing DiffReps. Log2 fold-changes in peak densities for H3K27Ac, BRD4, P300, and ATAC are denoted by blue bars.