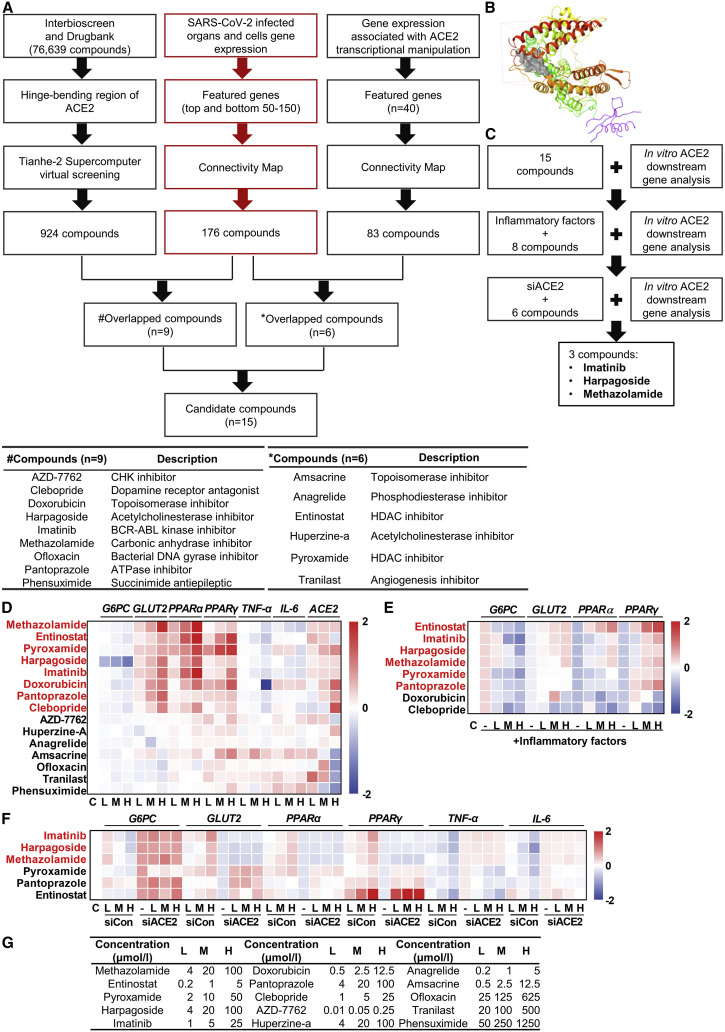
Figure 3.
Imatinib, harpagoside, and methazolamide are identified as ACE2 activators
(A) Flow chart depicting the process of in silico identification of potential ACE2 activators with Tianhe-2 supercomputer virtual screening and CMAP bioinformatic analysis (top) and detailed information of the overlapped compounds (bottom, # and ∗ indicated different overlapping strategy).
(B) The docking site (gray) of ACE2 protein (PDB: 1R42).
(C) Schematic of the in vitro experimental workflow to identify the final three compounds.
(D–F) Real-time PCR of gene expression in HUVECs. Log10 fold change was calculated based on treated samples against control samples. HUVECs were treated with 15 compounds for 16 h (D), inflammatory factors for 32 h (50 ng/mL TNF-α, IL-4, IL-6, and IFN-γ) following eight compounds for 16 h (E), or 10 nM control siRNA (siCon) and ACE2 siRNA (siACE2) for 8 h following six compounds for 16 h (F) (n = 3). The selected compounds based on the scoring system were highlighted in red and applied for the following screening.
(G) The low (L), medium (M), and high (H) concentrations of 15 compounds applied in the above three experiments.
See also Figure S3.