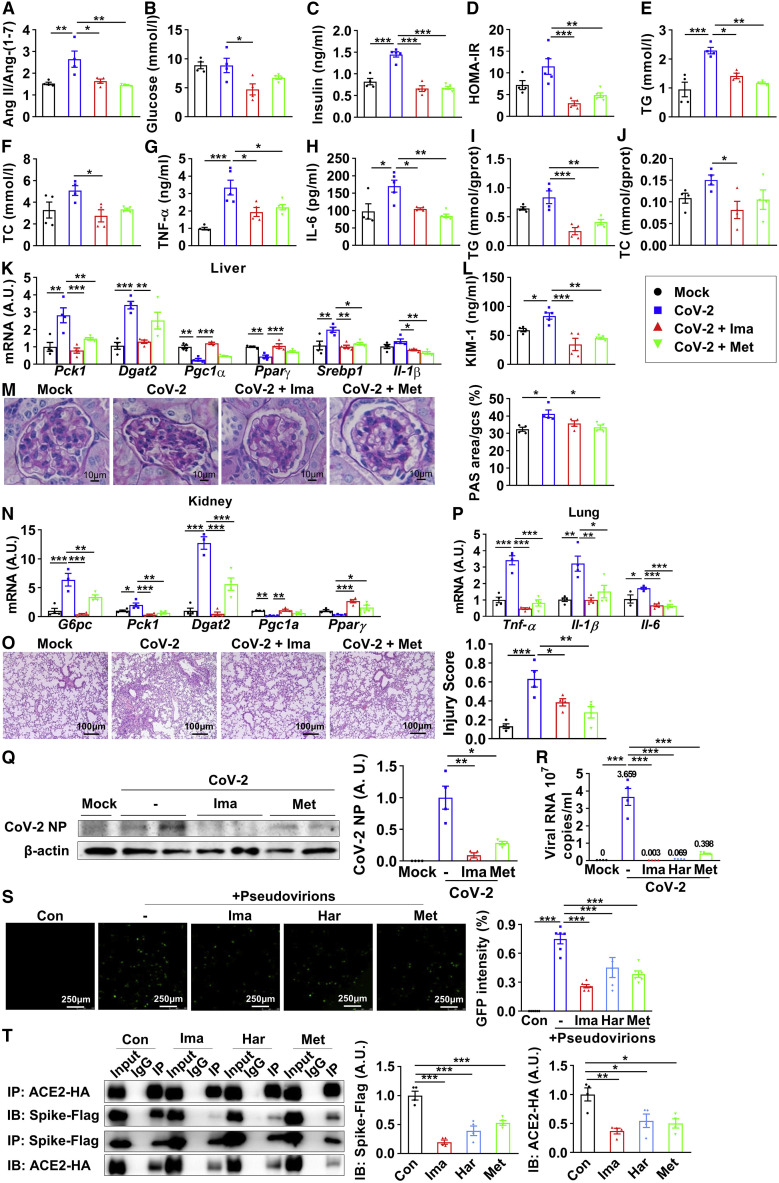
Figure 6.
ACE2 enzymatic activators improve metabolic defects and inhibit virus entry upon SARS-CoV-2 infection
(A–Q) Twelve-week-old human ACE2 transgenic mice were given vehicle (Mock, CoV-2), 250 mg/kg imatinib (CoV-2 + Ima), and 100 mg/kg methazolamide (CoV-2 + Met) through gavage once each day for 4 weeks after 6-week high-fat-diet treatment and were intranasally challenged with 4 × 104 FFU SARS-CoV-2. After 7 days of infection, all mice were fasted for 6 h and sacrificed.
(A–H) Quantification of the ratio of Ang II against Ang-(1–7) in plasma (A), fasting blood glucose (B), plasma insulin (C), homeostatic model assessment of insulin resistance (HOMA-IR) (D), plasma triglyceride (E), plasma total cholesterol (F), plasma TNF-α (G), and plasma IL-6 (H) (n = 4).
(I–K) Livers were subjected to lipid assay (I and J) and real-time PCR (K) (n = 3–4).
(L) Plasma KIM-1 (n = 4–5).
(M and N) Kidneys were subjected to periodic acid-Schiff (PAS) staining (M, images and quantification were shown; n = 4) and real-time PCR (N) (n = 4).
(O–Q) Lungs were subjected to H&E staining (O, images and quantification were shown; n = 4), real-time PCR (P) (n = 4), and immunoblotting of viral nucleocapsid protein (NP) (Q, blot shown on the left, quantification on the right; n = 4).
(R) Vero E6 cells were pretreated with 25 μM imatinib, 100 μM harpagoside, or 100 μM methazolamide for 6 h, followed by SARS-CoV-2 (MOI = 0.005) infection for 42 h; the supernatant was subjected to real-time PCR (n = 4).
(S) HEK293T cells expressing hACE2 were pretreated with 25 μM imatinib, 100 μM harpagoside, or 100 μM methazolamide for 6 h, followed by pseudovirion treatment for 66 h, and were observed with fluorescence microscopy (images shown on the left, quantification on the right; n = 5).
(T) The HEK293T cells overexpressing spike-FLAG and ACE2-HA were treated with 25 μM imatinib, 100 μM harpagoside, or 100 μM methazolamide for 48 h. Precipitated proteins were subjected to immunoblotting (blot shown on the left, quantification on the right; n = 4).
Error bars represent SEM. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. See also Figure S6.