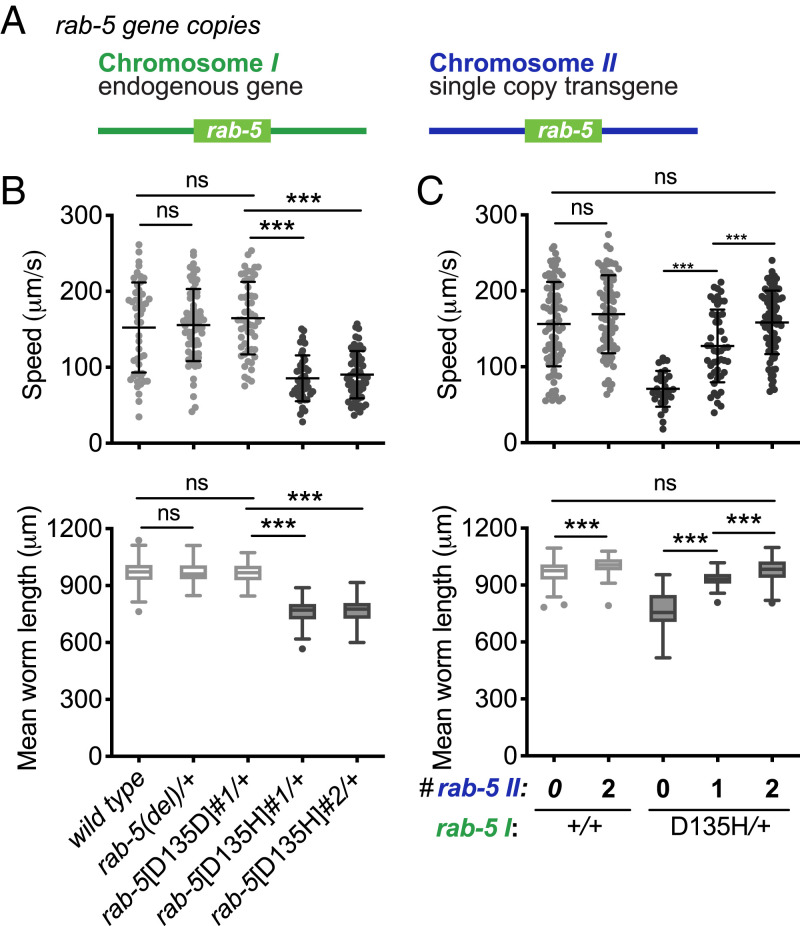
Fig. 2.
C. elegans rab-5[D135H] produces a dominant negative RAB-5 small GTPase. (A) Illustration of the endogenous rab-5 locus on chromosome I and the single-copy genomic wild-type rab-5 transgene integrated into a safe harbor locus on chromosome II. (B) Mean locomotion speed and worm length on growth media agar plates for the wild-type, rab-5(del) heterozygotes, rab-5[D135D]#1 control edit heterozygotes, rab-5[D135H]#1 heterozygotes, and rab-5[D135H]#2 heterozygotes (tested animals were cross-progeny from mating). (C) Locomotion speed and mean worm length on growth media agar plates. The wild type and the wild type homozygous for the single wild-type copy insertion of rab-5 on chromosome II (light gray) were self-progeny, while rab-5[D135H]#2 heterozygotes with the normal version of chromosome II hemizygous or homozygous for the chromosome II containing a single wild-type copy of rab-5 (dark gray) were cross-progeny from mating. Allele designations for independent edits (#1, #2) are in SI Appendix, Table S2. Three independent biological replicates were combined for each genotype. n ≥ 46 per condition, except for rab-5[D135H]#2 heterozygotes in C (n = 29). Scatterplots showing mean and SD are presented for locomotion speed. Filled circles indicate the average speed per animal for up to 1 min. Box plots indicate the mean and first and third quartiles, and whiskers indicate the 5th and 95th percentiles for measures of animal length. Differences between groups were determined by the Student’s t test. ns, not significant. ***P < 0.001.