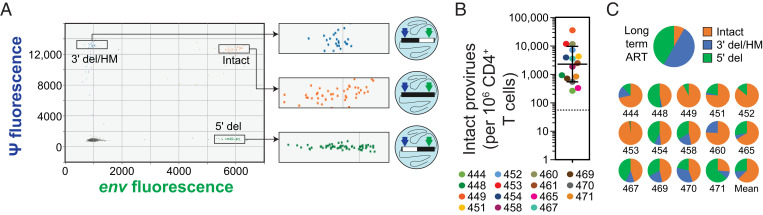
Fig. 2.
IPDA analysis of CD4+ T cells from PLWH prior to ART initiation. (A) Representative baseline (pre-ART) IPDA dot plot showing fluorescent signal from the packaging signal (ψ) amplicon at the 5′ end of the genome (y axis) and the env amplicon at the 3′ end of the genome (x axis) for individual proviruses suspended in nanoliter-sized droplets (illustrated in diagrams to the Right of each expanded box). Proviruses with 5′ deletions (5′ del) appear in the lower right quadrant. Proviruses with 3′ deletions and/or APOBEC3-mediated hypermutation (3′ del/HM) appear in the upper left quadrant. Intact proviruses are directly counted as double-positive droplets in the upper right quadrant. Raw droplet counts are corrected for shearing between the amplicons. For assay details, see ref. 41. (B) Baseline frequency of intact proviruses in circulating CD4+ T cells prior to initiation of ART. Solid lines, geometric mean ± SD; dotted line, median frequency of intact proviruses among 400 PLWH on long-term ART for comparison, based on ref. 43. Baseline samples were not available for participants 441 and 447. (C) Mean fraction of intact, 5′ del, and 3′ del/HM proviruses in CD4+ T cells from PLWH. (C, Upper) Mean fractions from 400 PLWH on long-term ART (for comparison, based on ref. 43). (C, Lower) Fractions for individual participants in this study prior to ART. Baseline samples were not available for participants 441 and 447. For participant 461, defective proviruses could not be detected above the background level of single positive droplets generated by DNA shearing.