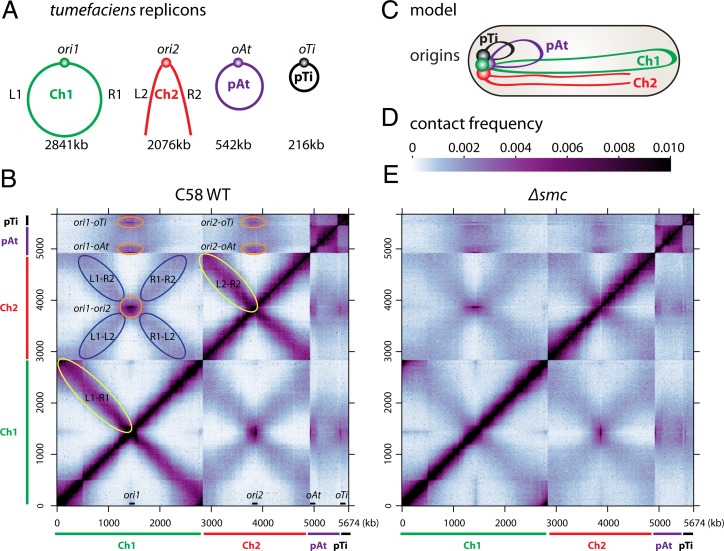
Fig. 1.
Genome-wide organization of A. tumefaciens replicons. (A) A. tumefaciens C58 wild-type cells have a circular chromosome (Ch1), a linear chromosome (Ch2), and two plasmids (pAt and pTi). The replication origins are labeled ori1, ori2, oAt, and oTi. The left and right arms of the chromosomes are labeled L1, R1, L2, and R2. The sizes of these four replicons are indicated. (B) Normalized Hi-C contact map displaying contact frequencies for pairs of 10-kb bins across the genome of A. tumefaciens C58 wild-type (WT). The x and y axes indicate genome positions. To better visualize contacts in the origin region of Ch1, the reference genome of Ch1 is rearranged with the origin (ori1) at the center and the two arms on either side. Ch1, Ch2, pAt, and pTi are indicated by green, red, purple, and black bars, respectively. The positions of the four origins are indicated on the x axis. Interarm interactions on both Ch1 and Ch2 are circled in yellow. Interactions between origins are circled in orange. The interactions between the arms of Ch1 and the arms of Ch2 are circled in blue. (C) Schematic model of genome organization in a newborn cell. The four origins are clustered together at the old cell pole. Ch1 and Ch2 are aligned along the cell length. (D) The scale depicts Hi-C interaction scores (contact frequency) for all contact maps presented in this study. (E) Normalized Hi-C contact map of Δsmc (AtWX108). Interarm interactions are absent in Ch1 and Ch2.