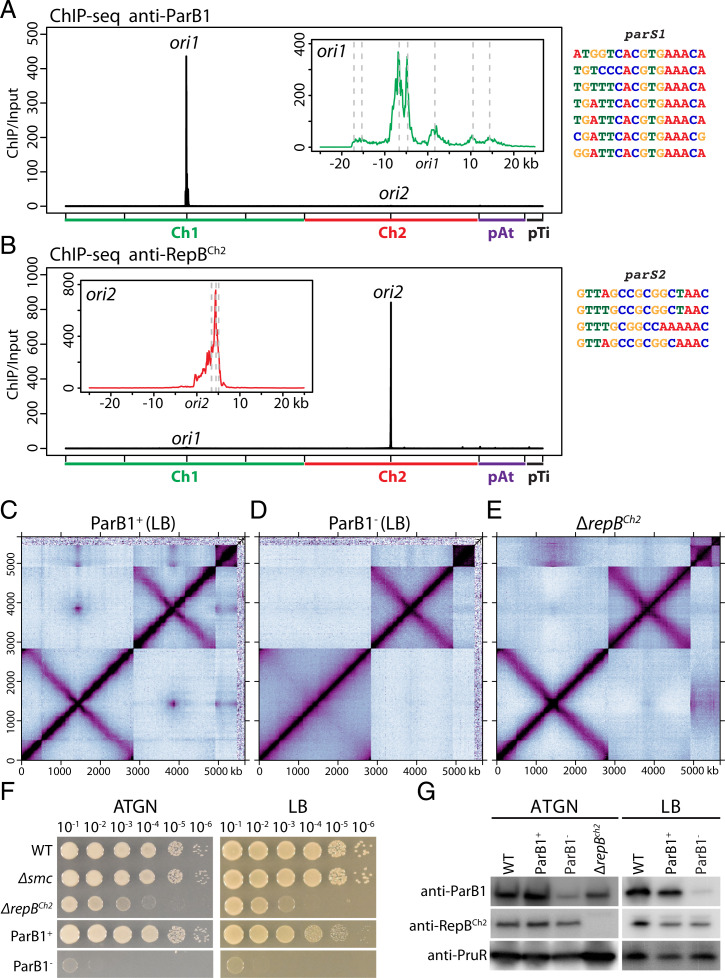
Fig. 2.
SMC loaders on Ch1 and Ch2. (A) Left: ParB1 enrichment in wild-type cells. To match the presentation of Hi-C, the reference genome of Ch1 is rearranged with the origin (ori1) at the center and the two arms on either side. Sequencing reads from ChIP and input samples were normalized to the total number of reads. The x axis shows genome positions, and the y axis indicates ChIP enrichment (ChIP/input) in1-kb bins. Inset: High-resolution plots of a 50-kb region encompassing ori1. Putative parS1 sites are indicated by gray dotted lines. Data are plotted in 100-bp bins. Right: Individual parS1 sequences are aligned. (B) Left: RepBCh2 enrichment in wild-type cells. Inset: High-resolution plots of a 50-kb region encompassing ori2. Putative parS2 sites are indicated by gray dotted lines. Right: Individual parS2 sequences are aligned. (C and D) Normalized Hi-C contact maps for ParB1 depletion strain (AtWX192) grown in LB with (ParB1+) or without (ParB1−) AHL and theophylline. A different genetic isolate is shown in SI Appendix, Fig. S4 K and L. 30-h depletion was used for this experiment. An earlier time point (12-h depletion) is shown in SI Appendix, Fig. S4N. The same strains growing in ATGN were shown in SI Appendix, Fig. S4 E–H. When ParB1 was depleted, interarm interactions are absent on Ch1 but unchanged on Ch2. Quantification of the interactions can be found in SI Appendix, Fig. S5. We observed the loss of pTi in this strain in two genetic isolates (SI Appendix, Fig. S4 E–L), likely during strain construction. (E) Normalized Hi-C contact maps for ΔrepBCh2 (AtWX089). When repBCh2 was deleted, interarm interactions on both chromosomes were unchanged. (F) Tenfold serial dilutions of the indicated strains spotted on ATGN plate (left) or LB plate (right). ParB1+ plates contain 1 μM AHL and 2 mM theophylline, and ParB1− plates lack these additives. (G) Immunoblot analysis showing levels of ParB1, RepBCh2, and PruR, a loading control. Wild-type, ParB1 depletion strain (AtWX192), and ΔrepBCh2 (AtWX089) were grown in LB or ATGN as indicated. After 30-h depletion, ParB1 has ∼3% remaining in LB (also SI Appendix, Fig. S4M) and 21.0% in ATGN.