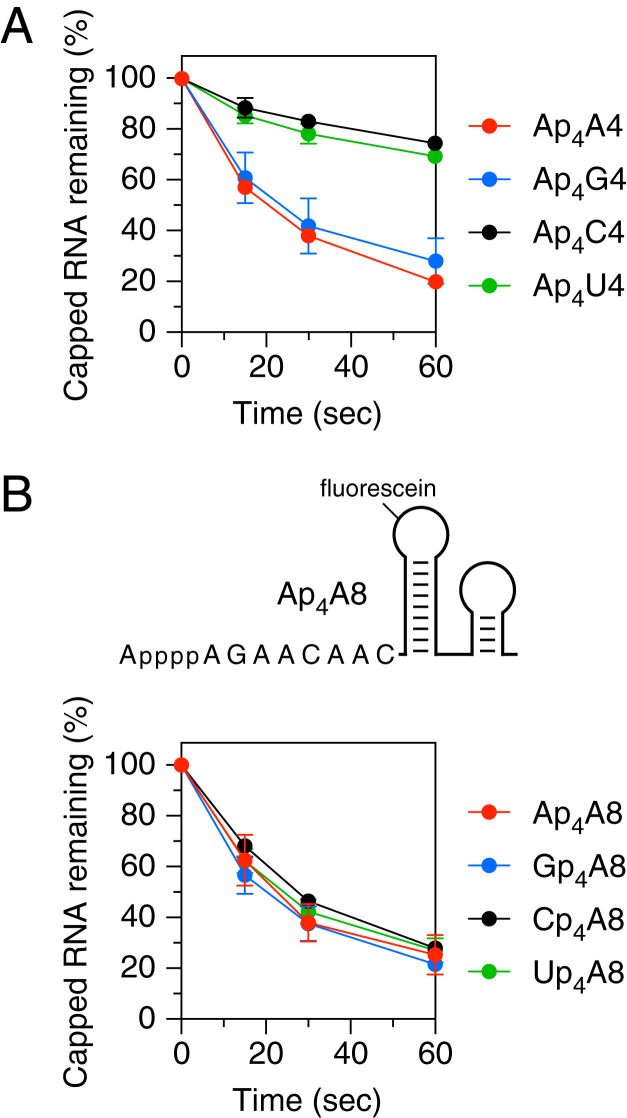
Fig. 3.
Effect of nucleotide identity on decapping by RppH in vitro. (A) Reactivity of substrates differing in the first transcribed nucleotide. The decapping of Ap4A4, Ap4G4, Ap4C4, and Ap4U4 by RppH was monitored as in Fig. 2. (B) Reactivity of substrates differing in the cap nucleotide. (Top) 5′-terminal sequence and expected secondary structure of Ap4A8. (Bottom) Decapping of Ap4A8, Gp4A8, Cp4A8, and Up4A8 by RppH. The 5′-terminal sequences of these substrates are shown in SI Appendix, Fig. S1. Corresponding time courses for decapping of the internal standard Ap4A8XL by RppH are graphed in SI Appendix, Fig. S2 B and C. Each time point is the average of three or more independent measurements. Error bars correspond to SDs.