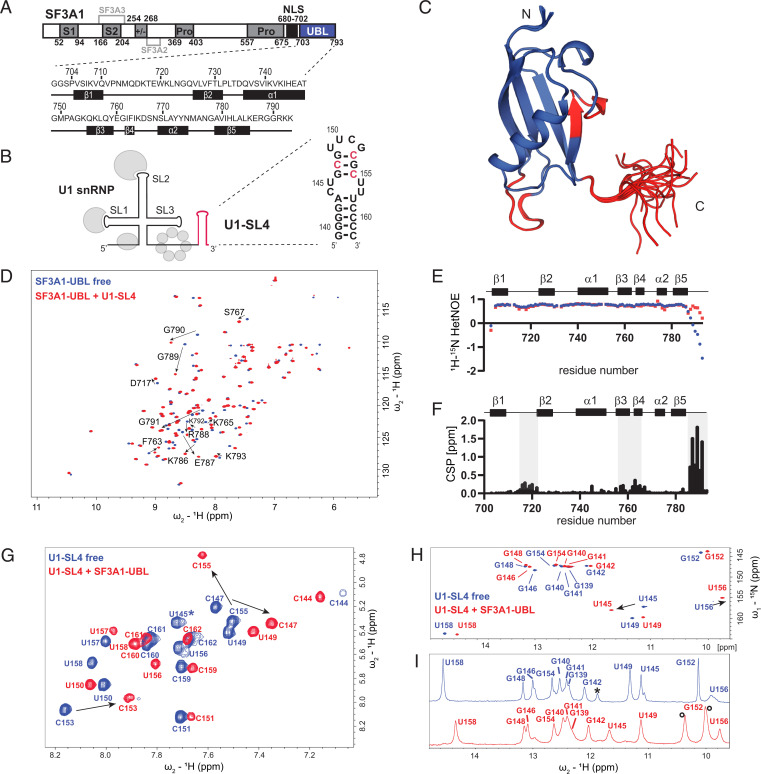
Fig. 1.
Interaction of SF3A1-UBL with U1-SL4. (A) Domain organization of SF3A1 and primary sequences of the SF3A1-UBL construct used in this study, including the secondary structure elements shown below. S1 and S2, SURP1 and SURP2; Pro, proline-rich sequence; +/−, charged residues; NLS, nuclear localization signal. Interaction sites for SF3A2 and SF3A3 are highlighted in light gray. (B) Schematic representation of U1 snRNP with U1-SL4 shown in red and the predicted secondary structure of U1-SL4 on the right. Nucleotides with the strongest CSP in 2D 1H-1H TOCSY are shown in red. (C) Solution structure of the free SF3A1-UBL. Overlay of the 20 lowest energy structures is shown. Amide CSPs of D and F shown in red. (D) Overlay of 2D 1H-15N heteronuclear single quantum coherence (HSQC) spectra of 15N-labeled SF3A1-UBL in the free (blue) and U1-SL4 RNA-bound (red) 1:1 complex form. CSPs of C-terminal residues are indicated with black arrows. (E) Backbone dynamics data of SF3A1-UBL in the free (blue) and U1-SL4 bound states (red). (F) Plot of the combined chemical shift difference between amide group resonances of the free and bound forms of SF3A1-UBL. (G) Overlay of 2D 1H-1H TOCSY spectra of U1-SL4 free (blue) and bound to SF3A1-UBL (red). Asterisk indicates U145, which was not identified in the bound state in 2D 1H-1H TOCSY. (H and I) Overlay of 2D 1H-15N HSQC and 1D 1H spectra, respectively, of imino signals of U1-SL4 free (blue) and bound to SF3A1-UBL (red). Black asterisk in I indicates an imino signal deriving from a duplex conformation. Black circles in I indicate protein amide signals.