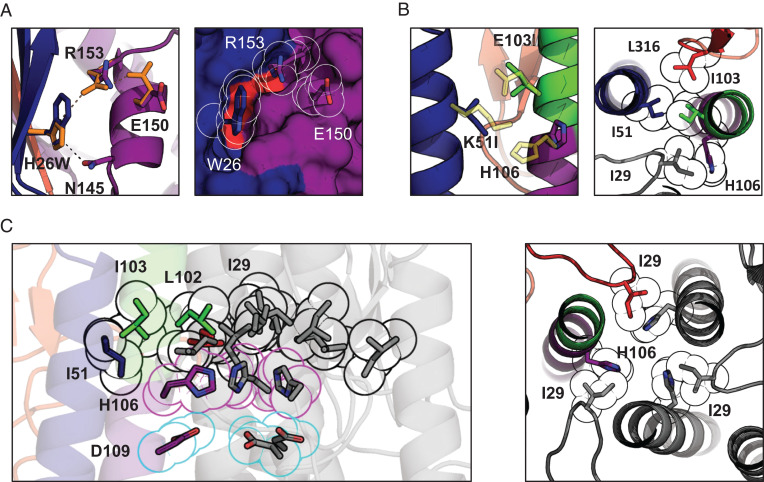
Fig. 3.
Crystal structure and suggested mechanism of action of the HA-stabilizing mutations. (A) Changes in the sidechain orientation induced by the H26W mutation in pHS1 (Left). The structure of the WT pHS1 is plotted in orange. The structure of the mutant is colored according to the color scheme introduced in Fig. 1A. The mutated pHS1 is also plotted in space-filling representation (Right). W26 sidechain tightly packs between the β-sheet (blue) and R153. (B) Changes in the sidechain orientation induced by the K51I mutation in pHS2 (Left). The structure of the WT pHS2 is plotted in yellow. The structure of the mutant is colored according to the color scheme introduced in Fig. 1A. pHS2 together with two neighboring residues I103 and I29 is also shown in a top cross-section view (Right). All four aliphatic residues form a tight hydrophobic cluster with H106 turning away. (C) The hydrophobic cluster from B is shown for the whole trimer (black outline), with the other two protomers colored in gray. The cluster encompasses the three H106 residues facing each other (magenta outline) and stabilized by negatively charged D109 residues (cyan outline). The histidines form most tight interactions with I29 (Right), shown in cross-section perpendicular to the threefold axis from the top.