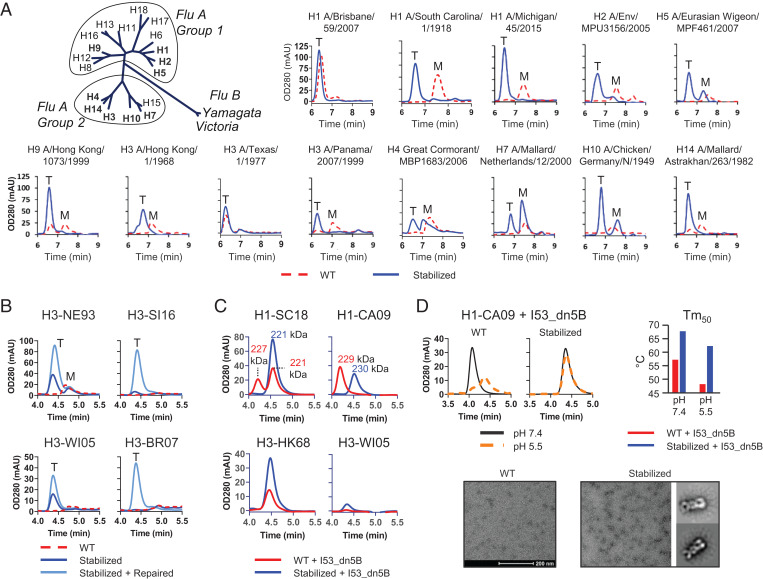
Fig. 4.
Stabilizing mutations applied to group 1 and 2 representative HAs. (A) Phylogeny of influenza A and B adapted from ref. 51; maximum likelihood tree representing amino acid sequences of HA. Analytical SEC profiles of culture supernatant of Expi293F cells expressing selected group 1 and 2 WT and fully stabilized HA (H26W, K15I, and E103I) subtype HAs. Peaks representing monomeric and trimeric species are indicated by an “M” and “T,” respectively. Due to differences in experimental setup, retention times are different from B, C, and D. (B) Analytical SEC profiles of culture supernatant of Expi293F cells expressing WT, stabilized and repaired of selected H3 HAs; H3N2 A/Netherlands/179/1993 (H3-NE93), H3N2 A/Singapore/INFIMH/16/0019/2016 (H3-SI16), H3N2 A/Wisconsin/67/2005 (H3-WI05), and H3N2 A/Brisbane/10/2007 (H3-BR07). (C) Analytical SEC profiles of Expi293F cell culture supernatants of H1N1 A/South Carolina/1/1918 I53_dn5B (H1-SC18), H1-CA09-I53_dn5B, H3-HK68-I53_dn5B, and H3-WI05-I53_dn5B. For the H1 HAs, the molecular weight determined by MALS is shown. (D) Analytical SEC profiles of purified WT and stabilized H1-CA09 HA-I53_dn5B kept at pH 5.5 and 7.4 for 2 wk (Top Left). Temperature stability as determined by DSF for WT and stabilized (Top Right). Negative-stained EM micrograph of WT and stabilized (Bottom) HA-I53_dn5B fusions. Representative 2D-averaged classes (box is 35 nm) could be generated for the stabilized variant.