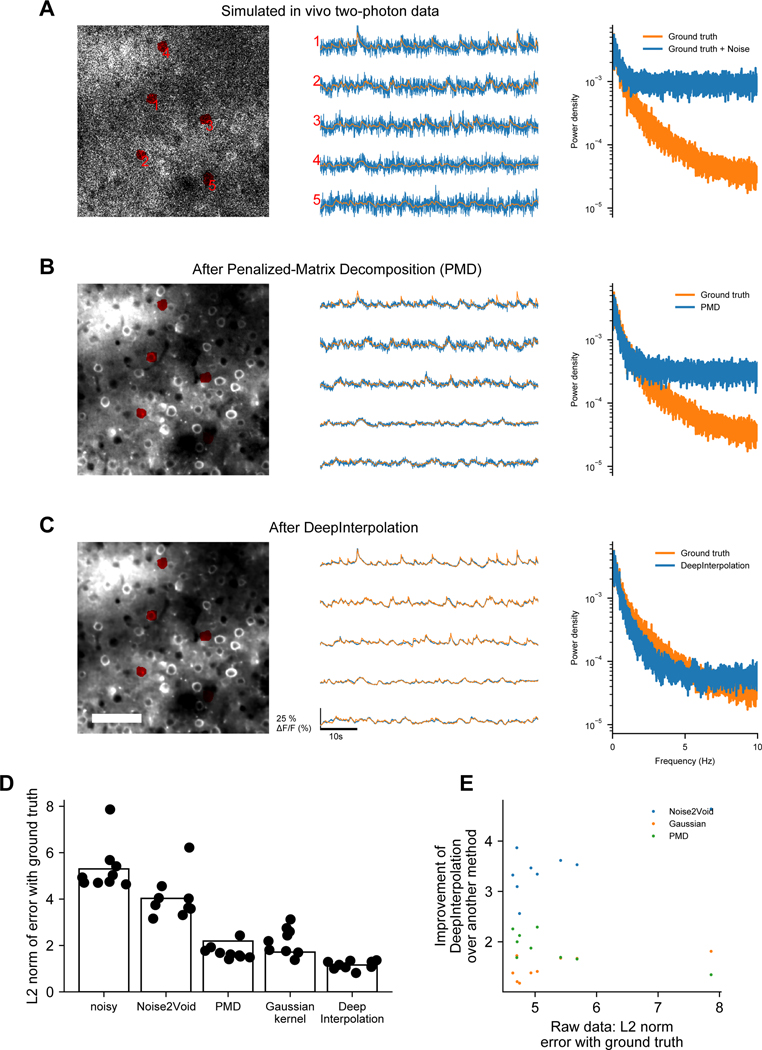
Extended Data Fig. 1. Comparison of DeepInterpolation, Noise2Void6, a Gaussian kernel, Penalized Matrix Decomposition (PMD)2on simulated calcium movies with ground truth.
(A) A simulated two photon calcium movie that was generated using an in silico Neural Anatomy and Optical Microscopy (NAOMi) simulation12. Left: A few ROI were manually drawn to extract the associated traces (middle) of the simulated movie with (blue trace) and without sources of noise (ground truth, orange). Right: The corresponding Fourier transform of all traces was calculated for each trace and averaged across. (B) The same movie and traces after PMD denoising. (C) The same movie and traces after DeepInterpolation. Scale bar is 100 μm. (D) Quantification of average mean squared error (L2) between noisy traces and ground truth, as well as between Noise2Void, PMD, a gaussian kernel (with σ=3) and DeepInterpolation with ground truth. (E) Quantification of the improvement in L2 norm against the original noisy error.