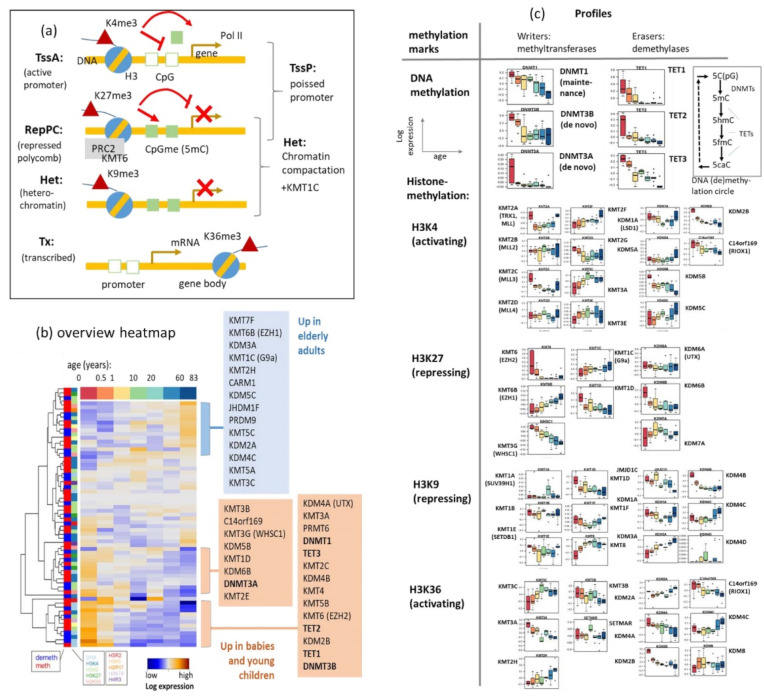
Figure 6.
Aging profiles of gene expression of a series of enzymes acting as writers or erasers of methylation marks at DNA and histone side chains. (a) Schematic sketch of the joint effect of histone lysine side chain and DNA methylation on the transcription of downstream genes and their relation to different chromatin states. (b) The overview heatmap shows that most of the genes, among them the DNA methyltransferases DNMT1 and DNMT3A,B and the demethylases Tet1–Tet3 (acting along the cytosine oxidation cycle) show large expression in babies and young children, which then decays with age, while another, smaller group gains expression upon aging, among them the H3K27me3 writing and chromatin compacting enzyme G9a (KMT1c). (c) Boxplots of the aging profiles of part of the enzymes reveal a variety of courses either increasing or decreasing with age. The different marks can have activating (e.g., triple methylation of lysine side chain H3K4 or H3K36), repressing (e.g., triple methylation at H3K27 or at H3K9) or poising (combination of H3K4 and H3K27) and chromatin compacting (H3K27 and H3K9) effect on transcription of the associated gene as schematically illustrated in part c. Combination of H3K4me3 and H3K27me3 gives rise to poised (“ready to go” and/plastic state) transcription while combination of H3K27me3 and H3K9me3 leads to compacted chromatin. For a detailed description of the enzymes and the histone code, see [24,68,69,70,71] and [66,67,72] for mechanisms in brain development. Maps of the different enzyme classes in the expression landscape of the brain are shown in Figure A7. DNA cytosine (CpG) methylation and de-methylation oxidation cycle are shown in part b.