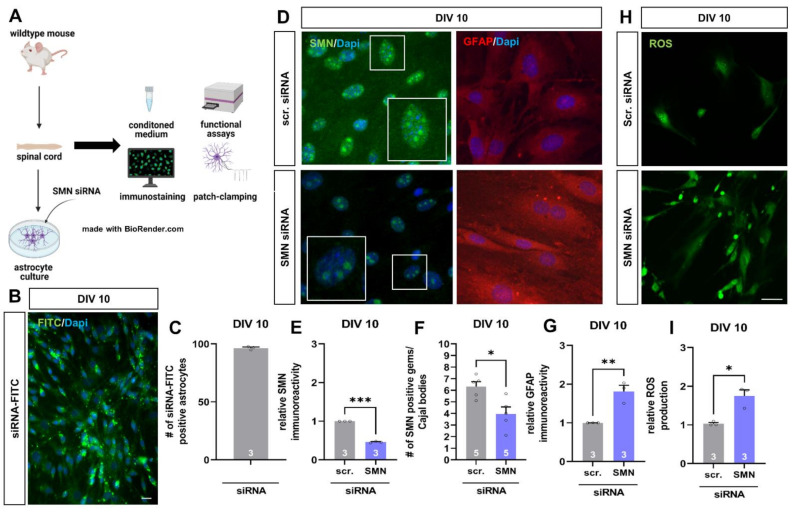
Figure 3.
SMN-deficient spinal astrocytes can be generated by SMN siRNA in vitro, showing similar protein alteration as spinal astrocytes in the late-onset SMA mouse model. (A) Schematic drawing of the generation of SMA-deficient spinal astrocyte cultures by SMN siRNA and performed experiments. (B) Transfection of wild-type astrocyte cultures with scrambled siRNA-FITC (green) by magnetic transfection method. Nuclear DNA was stained with Dapi (blue). (C) Cultured wild-type astrocytes were successfully transfected with siRNA-FITC using the magnetic transfection method. (D) Immunostaining of wild-type astrocyte cultures transfected with scrambled (scr.) or SMN siRNA against SMN protein (green) and GFAP (red) at DIV 10. In addition, nuclear DNA was stained with Dapi (blue). (E) Transfection of wild-type spinal astrocytes with SMN siRNA resulted in a reduction of SMN protein similar to the level observed in the juvenile SMA type III mouse model (***, p < 0.001). (F) SMN positive gems/Cajal bodies in the nucleus of astrocytes were reduced by transfection with SMN siRNA (*, p < 0.05). Furthermore, an increased number of GFAP-positive aggregate-like structures were observed in SMN-deficient astrocytes (white arrows). (G) SMN-deficiency in spinal astrocytes induced by SMN siRNA resulted in increased GFAP level (**, p < 0.01). (H) Staining of ROS production (green) in wild-type astrocytes transfected with scrambled or SMN siRNA at DIV 10. (I) SMN-deficient astrocytes showed an increased production of ROS compared to astrocytes transfected with scrambled siRNA (*, p < 0.05). n = 3–5 independent experiments (see bars). For each experiment > 50 cells were analyzed. Scale bar: B = 50 µm; F = 20 µm.