Abstract
The circadian clock orchestrates an organism’s endogenous processes with environmental 24 h cycles. Redox homeostasis and the circadian clock regulate one another to negate the potential effects of our planet’s light/dark cycle on the generation of reactive oxygen species (ROS) and attain homeostasis. Selenoproteins are an important class of redox-related enzymes that have a selenocysteine residue in the active site. This study reports functional understanding of how environmental and endogenous circadian rhythms integrate to shape the selenoproteome in a model eukaryotic cell. We mined quantitative proteomic data for the 24 selenoproteins of the picoeukaryote Ostreococcus tauri across time series, under environmentally rhythmic entrained conditions of light/dark (LD) cycles, compared to constant circadian conditions of constant light (LL). We found an overrepresentation of selenoproteins among rhythmic proteins under LL, but an underrepresentation under LD conditions. Rhythmic selenoproteins under LL that reach peak abundance later in the day showed a greater relative amplitude of oscillations than those that peak early in the day. Under LD, amplitude did not correlate with peak phase; however, we identified high-amplitude selenium uptake rhythms under LD but not LL conditions. Selenium deprivation induced strong qualitative defects in clock gene expression under LD but not LL conditions. Overall, the clear conclusion is that the circadian and environmental cycles exert differential effects on the selenoproteome, and that the combination of the two enables homeostasis. Selenoproteins may therefore play an important role in the cellular response to reactive oxygen species that form as a consequence of the transitions between light and dark.
Keywords: selenocysteine, circadian clock, selenoproteome, selenium, cellular rhythms
1. Introduction
Circadian clocks have evolved in all kingdoms of life as an adaptation to the rhythmic environment on planet Earth. An organism’s circadian clock enables the anticipation of predictable daily changes in the environment [1]. When an organism is subjected to a rhythmic environment, such as light/dark cycles, the circadian clock orchestrates processes such as metabolism and photosynthesis to synchronise to the external cycles. Circadian regulation exists at all levels of cellular organisation, including the rhythmic expression of up to one-third of the transcriptome [2,3]. On top of circadian regulation, strong direct responses exist to all rhythmic environmental parameters, but in particular to light/dark cycles. Direct responses to environmental cycles can be disentangled from circadian effects by studies performed under constant environmental conditions.
In a eukaryotic cell, levels of reactive oxygen species (ROS) oscillate over the 24 h cycle, and are influenced by the circadian clock as well as by environmental light/dark cycles [4,5,6,7,8]. Oscillations in the cell redox state are created either from ROS production by rhythmic cellular metabolism (via the mitochondria or NADPH oxidases, or photosynthesis in the green lineage), or via the rhythmic expression of antioxidant proteins within the cell [5]. Powerful oxidising agents, ROS are small, short-lived molecules, involved in the regulation of multiple cell functions and pathways, and include superoxide (O2−) and hydrogen peroxide (H2O2). Excessive ROS accumulation can result in oxidative damage and increase cell toxicity, eventually leading to cell death. However, balanced fluctuations of reduction–oxidation (redox) homeostasis have important signalling functions [5,9], implying a tight regulation of redox over the 24 h cycle to compensate for, and integrate, the rhythmic environment.
One group of proteins predominantly involved in redox homeostasis are selenoproteins. First identified by Thressa Stadtman in 1973 [10], selenoproteins are proteins containing a selenocysteine residue. Known as the 21st amino acid, selenocysteine (Sec, or U) is not directly encoded for in the standard genetic code; instead, it is encoded by the opal stop codon UGA through translational recoding that relies on a highly specific cis-acting stem–loop structure in the 3′ UTR of the transcript (the selenocysteine insertion sequence; SECIS [11,12,13]). Many selenoproteins function as redox-related enzymes, and selenoenzymes confer a higher catalytic activity than enzymes with a normal cysteine in the active site: the selenol group is far more reactive than the thiol group in a cysteine amino acid, making selenoproteins highly physiologically active [11]. Selenoproteins play critical roles in regulating the cellular redox state and the prevention and repair of ROS-induced damage to cellular components [12,14]. Although selenoproteins are found within bacteria, archaea, and eukaryotes, many organisms have lost the machinery to utilise selenium to create selenocysteine [14], and have cysteine residues in the corresponding positions of enzymes.
Species of phytoplankton perform nearly half of the world’s photosynthesis, and play a critical role as primary producers in global food webs [15]. Ostreococcus tauri is one of the major species of marine phytoplankton—a eukaryotic green alga with a small, minimal genome of 12.6 million base pairs, made up of 20 haploid chromosomes [16]. The most recent gene models for this organism reveal that of the 7700 genes contained in its genome, 24 encode for selenoproteins [17]. Ostreococcus has a simple, plant-like transcriptional circadian clock [18], which is regulated at the post-translational and metabolic levels by processes shared between eukaryotes [19,20,21,22]. We previously published a detailed proteomic time series in Ostreococcus tauri under natural cycles of light and dark (LD; 12 h/12 h cycles of light/dark), as well as under constant light conditions (LL) [23]; that dataset covered all of the 24 Ostreococcus selenoproteins, allowing an unprecedented insight into environmental and circadian regulation of the selenoproteome in a eukaryotic model cell.
In this study, we assessed the rhythmicity of selenoproteins under environmental 24 h cycles of LD versus under circadian conditions of LL. We also investigated the effects of the light/dark cycle and the circadian clock on cellular selenium uptake and, reciprocally, the effect of selenium deprivation on the circadian clock. Our combined results point to complex interactions between endogenous circadian regulation and environmental light/dark cycles that together shape the temporal selenoproteome.
2. Materials and Methods
Proteomics data were mined from our publicly available previous study [23], and results from samples were collected every 3.5 h across one cycle of 12 h light/12 h dark (LD, or entrained conditions) and three cycles under constant light conditions (LL, or circadian conditions). Cells were grown as documented previously.
Selenoproteins were defined as proteins containing a selenocysteine (U). Selenoproteins were named using the gene function and protein domain information available on ORCAE [24], as well as the functions of homologous proteins in other model species. These protein homologs were determined by Domain Enhanced Lookup Time Accelerated Basic Local Alignment Search Tool (DELTA-BLAST) [25], using the amino acid sequence of the Ostreococcus protein. The DELTA-BLAST searches were initially limited to homologs in Homo sapiens, Mus musculus, and Drosophila melanogaster. If there were no closely related homologous proteins in these model species, the search was widened and the three most closely related homologs—which were generally algal species—were retained. To assist in determining the functions of these selenoproteins and their homologs, Gene Ontology (GO) codes associated with Ostreococcus selenoproteins and related homologs were taken from UniProt, and GO definitions were taken from QuickGO [26]. To determine whether any selenoproteins were targeted to the organelles within the alga, TargetP-2.0 was used to identify the sequences found before the N-termini of the proteins [27]. Signal peptides were denoted as thylakoid luminal transit peptide (luTP), chloroplast transit peptide (cTP), mitochondrial transit peptide (mTP), or secretory pathway (SP) [23].
The rhythmicity parameters of proteins in both LD and LL were calculated using the eJTK_cycle algorithm in BioDare2 [28], with linear detrending for LL data and without detrending for LD data, as fully explained in [23]. Phase and relative amplitude were calculated using MFOURFIT and the LL proteome period was calculated by MESA, as reported previously [23]. A full rationale for the analysis methods and evidence for the accuracy of the rhythmicity parameters for both the single LD cycle and 3 LL cycles were reported previously [23].
ICP-MS analyses for the selenium isotopes 78Se and 82Se were performed as previously described [29]. Luminescence data were collected on a TriStar 2 plate reader (Berthold Technologies, Bad Wildbad, Germany) under LD or LL conditions, using 2 µmol/m2/s of blue light (Moonlight Blue filter, Lee Lighting). The CCA1-LUC line is a translational fusion of the CCA1 protein with firefly luciferase, driven from the CCA1 promoter (pCCA1::CCA1-LUC), and this line was described elsewhere [18].
Statistics and graphs from the transcriptome and proteome data were calculated and plotted using GraphPad Prism Version 9.1.0. Unless otherwise stated, the statistical tests performed were nonparametric two-tailed Mann–Whitney U tests, with their significance indicated (ns = p > 0.05; * = p ≤ 0.05; ** = p ≤ 0.01; *** = p ≤ 0.001; **** = p ≤ 0.0001).
3. Results
3.1. The Selenoproteome over Diurnal and Circadian Cycles
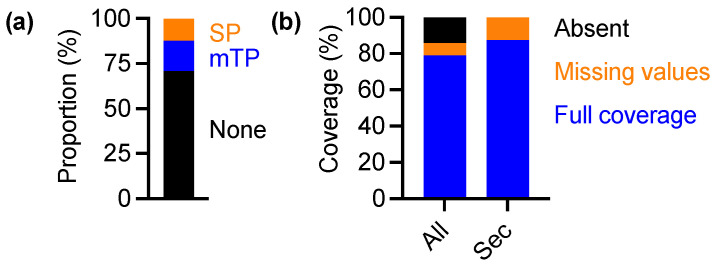
The nuclear genome of the green alga Ostreococcus tauri encodes 24 selenoproteins [17], while no predicted selenoproteins exist in the chloroplast or mitochondrial genomes [30]. We analysed the presence of predicted transit peptides at the N-termini of the selenoprotein sequences, and found that while a small number of proteins are predicted to be targeted to the mitochondrion (mitochondrial transit peptide; mTP) or secretory pathway (SP), none have chloroplast transit peptides (Figure 1a). The vast majority of selenoproteins have no previously described transit peptide (None); therefore, it is likely that most selenoproteins are localised in the cytoplasm, and all of them are translated there.
Figure 1.
Proteome coverage of the selenoproteome: (a) Proportion of selenoproteome predicted to have a transit peptide for the secretory pathway (SP), mitochondrion (mTP), or no transit peptide (None). (b) Coverage of proteins containing a selenocysteine (Sec) versus the whole proteome (All) in [23]. ‘Absent’ refers to proteins that were not quantified at any of the timepoints in either the LD or LL time series. ‘Missing values’ refers to proteins that were quantified in some but not all data points of the time series, while ‘Full coverage’ refers to those proteins that were reliably quantified at every timepoint throughout the LD and LL time series. We previously published detailed proteomic time series in Ostreococcus tauri under natural cycles of light and dark (LD; 12 h/12 h cycles of light/dark), as well as under constant light conditions (LL), sampling at 3.5 h intervals [23]. In that study, we obtained 85% coverage of the theoretical proteome and, interestingly, all 24 selenoproteins were detected in the dataset (Figure 1b and Supplementary Table S1). A total of 21 out of 24 selenoproteins had full coverage over the LD and LL time series (blue in Figure 1b), while 3 had missing data points (orange). Based on this exceptional coverage, we conclude that our dataset provides an unprecedented insight into rhythmic regulation of the selenoproteome.
3.2. Differential Regulation of Selenocysteine Usage by Environmental and Circadian Rhythms
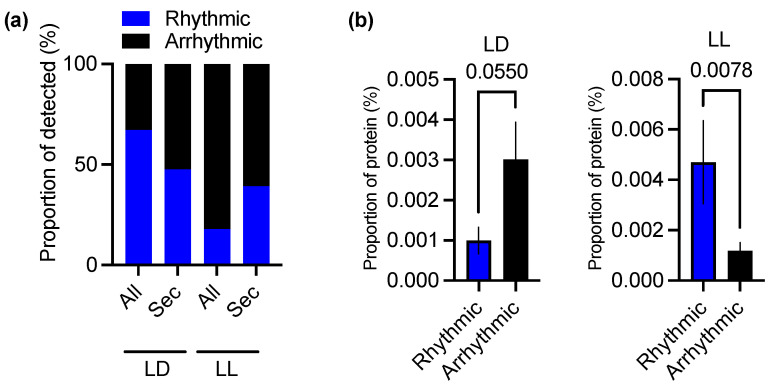
To gauge the overall level of rhythmic regulation of the selenoproteome, we compared the rhythmicity of selenoproteins versus the overall proteome. Rhythmicity of all protein abundance profiles was assessed using the eJTK algorithm [31,32], as described previously [23]. Abundance profiles over the time series can be found for all selenoproteins in Supplementary Figure S1. Compared to the overall Ostreococcus proteome, a lower proportion of selenoproteins were rhythmic under LD conditions (Figure 2a, Table 1). Surprisingly, the opposite was true under circadian conditions of constant light—a higher proportion of the selenoproteome was rhythmic compared to the overall proteome (Figure 2a).
Figure 2.
Rhythmicity of the selenoproteome: (a) Proportion of rhythmic versus arrhythmic proteins under LD or LL in the overall dataset reported in [23], or those proteins containing a selenocysteine (Sec). (b) Amino acid usage for selenocysteine, expressed as the average proportion of a protein amongst rhythmic or arrhythmic proteins under LD (left) or LL (right) conditions. Statistics are unpaired nonparametric tests (Mann–Whitney).
Table 1.
The Ostreococcus selenoproteins and rhythmicity parameters under light/dark cycles (LD) or constant light (LL): Provided are the p-value for rhythmicity, phase (in hours; h), relative amplitude (Rel. Amp., in percentage), and mean abundance (Mean Ab., in arbitrary units; a.u.) of the selenoproteins, as detected in our previous proteomics study [23]. ND = not done, meaning the analysis was not possible due to too many missing values in the dataset.
| Identifier | Description | LD | LL | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| p-Value | Phase (h) | Rel. Amp. (%) | Mean Ab. (a.u.) | p-Value | Period (h) | Phase (h) | Rel. Amp. (%) | Mean Ab. (a.u.) | ||
| ostta01g05530 | Thioredoxin-fold protein | 0.530 | 16.3 | 0.8 | 633,478 | 0.000 | 21.7 | 7.1 | 2.7 | 647,647 |
| ostta10g01410 | Selenoprotein F | 0.014 | 3.9 | 4.8 | 76,668 | 0.007 | 23.1 | 14.9 | 5.0 | 84,005 |
| ostta01g00700 | Disulphide isomerase 1 | 0.182 | 5.2 | 2.5 | 2,371,875 | 0.008 | 22.5 | 13.5 | 6.3 | 2,144,215 |
| ostta09g01390 | Selenoprotein U | 0.061 | 20.7 | 2.2 | 1,505,570 | 0.012 | 18.6 | 4.8 | 1.2 | 1,433,558 |
| ostta02g02950 | Selenoprotein H | 0.018 | 19.8 | 4.9 | 353,763 | 0.016 | 25.6 | 5.6 | 2.8 | 322,590 |
| ostta08g03450 | Glutathione peroxidase A | 0.000 | 6.8 | 6.3 | 2,441,906 | 0.027 | 21.6 | 15.8 | 7.4 | 4,076,098 |
| ostta09g00190 | Peroxiredoxin | 0.182 | 8.6 | 3.2 | 2,093,776 | 0.027 | 24.1 | 18.9 | 7.1 | 2,806,390 |
| ostta14g01560 | Selenoprotein K | ND | ND | ND | 41,402 | 0.031 | 25.7 | 9.4 | 1.9 | 41,354 |
| ostta01g06300 | Selenoprotein W | 0.123 | 9.3 | 5.9 | 574,635 | 0.039 | 24.2 | 17.8 | 10.6 | 889,748 |
| ostta09g00530 | Glutathione peroxidase C | 0.108 | 21.5 | 1.2 | 395,005 | 0.063 | 21.6 | 17.1 | 4.7 | 514,482 |
| ostta09g01720 | Methionine sulphoxide reductase A | ND | ND | ND | 1577 | 0.063 | 22.0 | 17.3 | 9.6 | 2771 |
| ostta12g02030 | Disulphide isomerase 2 | 0.000 | 5.3 | 7.3 | 503,473 | 0.102 | 22.2 | 5.9 | 3.8 | 377,407 |
| ostta02g02735 | Glutathione peroxidase E | 0.000 | 16.7 | 7.9 | 628,421 | 0.102 | 22.1 | 10.1 | 3.6 | 587,297 |
| ostta17g00710 | SAM-dependant methyltransferase | 0.037 | 20.5 | 1.4 | 485,420 | 0.125 | 21.3 | 4.8 | 2.1 | 487,887 |
| ostta01g04220 | Thioredoxin reductase | 0.008 | 13.8 | 4.5 | 951,929 | 0.219 | 24.4 | 1.6 | 2.5 | 1,150,422 |
| ostta08g03600 | Unknown | 0.000 | 22.0 | 6.8 | 258,959 | 0.281 | 25.8 | 3.7 | 3.2 | 262,933 |
| ostta18g01790 | Selenoprotein O | 0.016 | 23.6 | 5.4 | 328,866 | 0.281 | 23.3 | 11.1 | 0.6 | 379,395 |
| ostta05g01540 | Glutathione peroxidase B | 0.197 | 7.9 | 2.6 | 719,605 | 0.406 | 21.9 | 9.3 | 1.2 | 765,471 |
| ostta10g02090 | Membrane selenoprotein | 0.009 | 21.4 | 27.9 | 6724 | 0.656 | 27.6 | 2.7 | 10.0 | 6528 |
| ostta07g00300 | Glutathione peroxidase D | 0.061 | 16.5 | 1.5 | 552,907 | 0.688 | 34.2 | 23.3 | 0.6 | 612,140 |
| ostta13g00280 | Disulphide isomerase 3 | 0.669 | 13.9 | 0.7 | 604,624 | 0.750 | 23.4 | 8.4 | 2.2 | 601,203 |
| ostta10g00035 | Selenoprotein S | 0.106 | 22.5 | 10.7 | 144,751 | 0.781 | 21.8 | 16.4 | 4.0 | 137,022 |
| ostta03g04910 | Selenoprotein T | 0.106 | 16.5 | 1.8 | 113,777 | 0.844 | 17.8 | 3.8 | 1.2 | 114,652 |
| ostta04g01370 | Selenoprotein M | ND | ND | ND | 68,795 | ND | ND | ND | ND | 77,943 |
We next assessed the prevalence of the selenocysteine amino acid (Sec, or U) among rhythmic and arrhythmic proteins. As expected from the observed proportion of rhythmic selenoproteins, there was a significantly increased incidence of selenocysteines in rhythmic compared to arrhythmic proteins under LL conditions, while under LD conditions an opposite trend was observed that did not reach statistical significance (Figure 2b). The opposite trend for selenocysteine usage in rhythmic versus arrhythmic proteins under LD and LL conditions is remarkable, as overall the amino acid usage of the 20 standard amino acids is fairly consistent between LD and LL (Figure S2).
3.3. Phase Coordination of the Rhythmic Selenoproteome
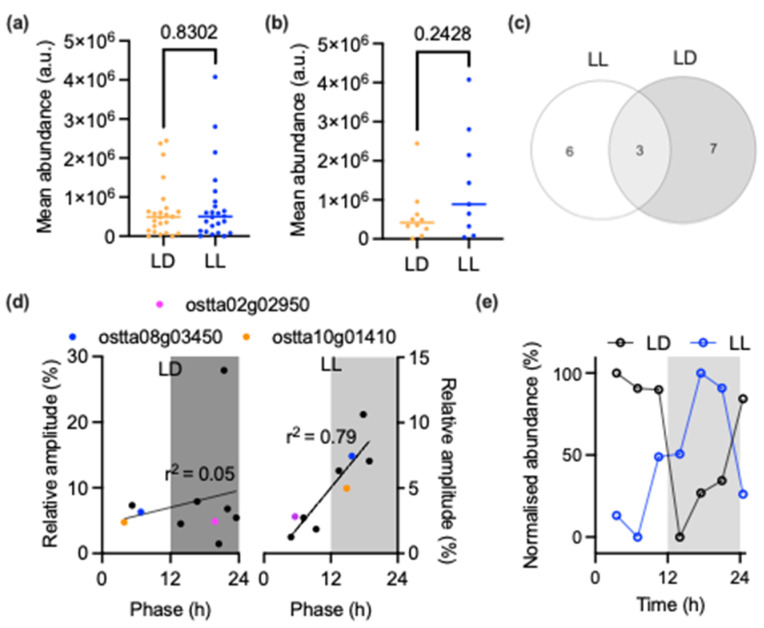
We then examined the circadian characteristics of the selenoproteins more closely. In accordance with the overall proteome [23], there was no difference in selenoprotein abundance between LD and LL conditions (Figure 3a); there was also no difference in relative amplitude between rhythmic selenoproteins (Figure 3b). Interestingly, this is a marked deviation from the trend in the overall proteome; generally, proteins oscillated with a far higher relative amplitude under LD cycles than under LL conditions [23]. Therefore, the light/dark cycle appears to exert less of an effect on the extent of selenoprotein rhythms than what would be expected based on the general proteome trends, while the circadian clock has a greater influence on selenoprotein abundance.
Figure 3.
Rhythmicity parameters of selenoproteins: (a) Mean abundance of all selenoproteins under LD versus LL conditions. (b) Mean abundance of selenoproteins classed as rhythmic under LD versus LL conditions. Statistics in (a,b) are unpaired nonparametric tests (Mann–Whitney); data are reported in arbitrary units (a.u.). (c) Overlap between rhythmic selenoproteins under LD versus LL conditions. (d) Peak abundance phase versus relative amplitude of rhythmic selenoproteins under LD (left) or LL (right) conditions. Coloured data points represent proteins that are arrhythmic under both conditions. Lines are linear regression with annotated r2 values. (e) Protein abundance across LD versus LL for ostta08g03450. The LD data reflect the single LD cycle, while the LL data are mean values across the three LL cycles in [23].
Only three selenoproteins were rhythmic under both conditions (Figure 3c), indicating that, similar to the overall proteome [23], selenoprotein rhythmicity is very different between LD and LL conditions. One of these three was a glutathione peroxidase (ostta08g03450), and two of them were thioredoxin-like proteins (selenoprotein H; ostta02g02950, and selenoprotein F; ostta10g01410) [33,34]. Interestingly, when time of peak phase for rhythmic selenoproteins was plotted against the amplitude of their rhythms, we observed that two of these proteins (ostta08g03450 and ostta10g01410) peak together, but at very different peak phases between the two conditions: near ZT6 in LD and CT16 in LL (Figure 3d). The third protein (ostta02g02950) followed this trend, but in the opposite direction to the other two. To indicate this effect more clearly, the protein abundance traces of the glutathione peroxidase are shown in Figure 3e, where the daytime peak under LD conditions and the subjective night peak under LL are evidently different.
When comparing the phase versus amplitude of all rhythmic selenoproteins under LD conditions, we observed no correlation between these two parameters (Figure 3d; r2 = 0.06); relative amplitude was consistent across rhythmic proteins regardless of phase. However, under constant light conditions there was a clear trend towards increased amplitude of oscillations in selenoproteins that peak at a later phase during the 24 h cycle (r2 = 0.79). Combined, the data in Figure 2 and Figure 3 suggest that rhythmicity within the selenoproteome is regulated by a complex integration of environmental light/dark cycles and endogenous rhythms.
3.4. Diurnal Transcript Abundance Rhythms Do Not Dictate Selenoprotein Abundance
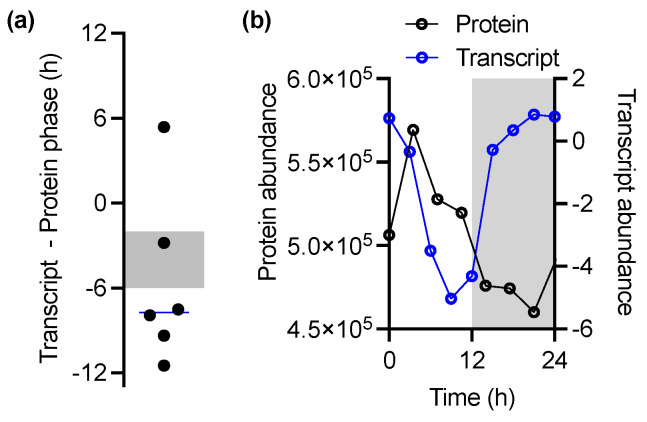
To test whether selenoprotein rhythmicity results from rhythmicity of the encoding transcripts, we compared selenoproteome profiles under LD cycles to publicly available transcriptomic data [35], in which we found 15 of the 24 selenoproteins (Supplementary Table S1). However, we did not observe a simple relationship between the timing of rhythmic transcripts and selenoproteins; for the six selenoproteins that were rhythmic at both the transcript and protein levels, only one protein peak phase lay between the 2–6 h after the transcript phase that would indicate a correlation [36,37], and the other five did not (Figure 4a, shaded box). An example is provided in Figure 4b, where the protein and transcript phase under LD cycles are close to antiphasic (ostta12g02030; disulphide isomerase)—transcript abundance shoots up directly following dusk, and remains high until the early morning, but this stark increase in transcript abundance after dusk is not followed by an increase in protein abundance until after dawn. Clearly, protein abundance depends not only on transcript abundance but also on the environmental light/dark cycle.
Figure 4.
No correlation between protein and transcript peak abundance phase: (a) One-for-one phase relationship between rhythmic selenoproteins and their cognate transcripts, expressed as the peak abundance phase of the transcript minus the peak abundance phase of the protein. Values that correspond to a protein that peaks in a 2–6 h window after its transcript would map to the shaded area. (b) Protein versus mean transcript abundance across the LD cycle for ostta12g02030.
3.5. Cellular Selenium Uptake Is Regulated by the Light/Dark Cycle
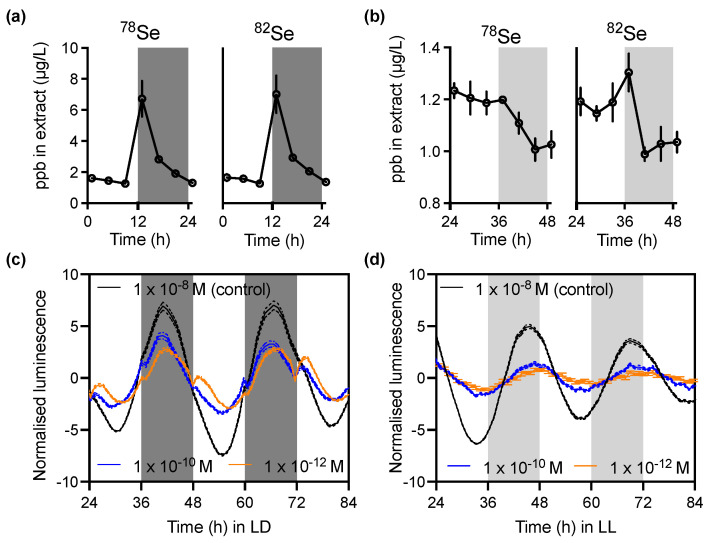
Selenoprotein synthesis requires an availability of selenium. To test whether instead of transcript abundance, selenoprotein abundance actually follows on from regulated selenium uptake by cells, we performed elemental composition analysis of cells. Inductively coupled plasma mass spectrometry (ICP-MS) was performed on whole-cell extracts to determine cellular selenium content under LD or LL conditions (Figure 5). Under LD conditions, the abundance of both observable isotopes of selenium was extremely rhythmic, with a sharp peak (nearly threefold increase) at the light-to-dark transition (Figure 5a); however, this uptake peak was almost entirely lost under constant circadian conditions (Figure 5b; note the different y-axis scales between panels a and b), indicating that selenium uptake rhythms are mediated through environmental signalling rather than endogenous circadian regulation. As selenium content was only elevated around dusk under LD conditions, and there was no apparent increased selenoprotein content at that time, we conclude that selenium uptake rhythms do not underlie selenoprotein rhythmicity.
Figure 5.
Interaction of selenium with the environmental and circadian cycles: (a,b) Cellular content of selenium, analysed by inductively coupled plasma mass spectrometry. The two detectable stable isotopes of selenium are 78Se (left) and 82Se (right), and these were measured under LD (a) or LL (b) conditions. Mean ± SEM; n = 3. (c,d) Luminescent imaging of the CCA1-LUC circadian clock marker under normal physiological concentrations of extracellular selenium (control) versus depleted levels 2 or 4 orders of magnitude lower. Cells were imaged over multiple cycles of 12 h/12 h light/dark (LD, (c)) or constant light (LL, (d)). Mean ± SEM; n = 32.
3.6. Functional Effects of Selenium Deprivation on Cellular Timekeeping
Finally, as we identified differential effects of light/dark cycles or constant circadian conditions on the abundance of selenoproteins as well as on selenium uptake, we tested the effects of selenium deprivation on circadian clock gene expression under both conditions. Cells were grown with normal concentrations of selenium (1 × 10−8 M, provided in the media as selenious acid; H2SeO3) or at 2 or 4 orders of magnitude lower. Clock gene expression was tested by longitudinal luminescent imaging of the circadian clock protein CCA1 translationally fused to the firefly luciferase enzyme [18]. Under LD conditions, selenium deprivation led to decreased amplitude of CCA1 expression rhythms (Figure 5c). More pronounced, however, selenium deprivation led to perturbed dynamics of clock gene expression around the light/dark transitions—at dusk, and especially at dawn, selenium-deprived cells responded with a large shoulder of CCA1 expression, while the control cells were not greatly affected by the light/dark transitions. Under constant circadian conditions, the lower amplitude in selenium-deprived cultures was consistent with LD, but the traces were qualitatively identical, without major period or phase defects (Figure 5d). Combined, this could imply that selenoproteins are important for the anticipation of—and compensation against—the effects of the light/dark cycle on timekeeping.
4. Discussion
We observed a limited similarity between rhythmicity of the selenoproteome under environmentally rhythmic conditions and circadian conditions of constant light—the identity of rhythmic selenoproteins and their peak abundance phases were different. However, in the absence of environmental rhythms, it is clear that the circadian clock shapes the selenoproteome over time; selenoproteins are overrepresented among rhythmic proteins under constant light conditions (Figure 2), and we observed a clear association between abundance of selenoproteins and circadian phase (Figure 3). This indicates a coordinated upregulation of these redox-related enzymes throughout the day. Under environmentally rhythmic conditions, on the other hand, the peak abundance of selenoproteins is spread evenly throughout the 24 h cycle, implying that the circadian regulation observed under LL conditions integrates with environmental signalling. This might mean that the circadian regulation that can be observed under constant light conditions exists merely to anticipate and counteract the effects of the environmental rhythms to enable homeostasis. Clearly, the selenoproteome is shaped by environmental cycles in combination with anticipatory effects achieved by circadian timekeeping.
A lack of correlation was observed between selenoprotein rhythmicity and their cognate transcript rhythms. This indicates that the rhythmicity of selenoproteins is defined by post-transcriptional processes. A clear shortcoming of this conclusion is that no transcriptome is available under constant circadian conditions for Ostreococcus, so analyses are limited to LD conditions. However, consistent with an overall proteome analysis [23], we conclude that selenoprotein abundance rhythms cannot be assumed from transcript data alone, nor can the rhythmicity of the protein under physiological rhythmic conditions be inferred from rhythmicity under constant circadian conditions.
The translation of selenoproteins requires selenium ions, which are lowly but sufficiently abundant in the sea water in which Ostreococcus grows. We pursued the hypothesis that rhythmically regulated selenium uptake might be the cause of increased prevalence of selenoproteins among clock-regulated proteins; however, the opposite was true—under circadian conditions we found no high-amplitude rhythms of cellular selenium content, whereas a clear diurnal pattern of selenium uptake was observed under light/dark cycles, with a peak directly following dusk. As this peak was largely absent under constant circadian conditions, it must result from a direct effect of darkness. Under light/dark cycles, however, selenoproteins are less likely to be rhythmic compared to the overall proteome, indicating that selenium uptake rhythms do not translate linearly to strong selenoprotein abundance rhythms. However, this result highlights the fact that selenium uptake is one of the effects of environmental rhythms that might integrate with circadian effects to influence the selenoproteome. Similarly, we found that selenium deprivation strongly affects the dynamics of circadian clock protein CCA1 under diurnal cycles, but not circadian conditions. When cells are grown in the absence of selenium, translation of selenoproteins terminates at the UGA codon, resulting in a truncated, non-functional enzyme [13]. It is therefore reasonable to assume that selenium depletion leads to lower levels of selenoproteins. As selenoproteins are a major class of redox homeostasis enzymes, and light/dark transitions are known to cause fluctuations in redox, it is tempting to speculate that selenoprotein activity buffers circadian gene expression against the effects of the light/dark cycle on the cellular redox state. As with our earlier results, this would be consistent with the notion that circadian regulation of the selenoproteome enables homeostasis under physiologically normal conditions.
In eukaryotes, only the genomes of mammals, small animals, plasmodia, and green algae encode confirmed selenoproteins [14]. This association with primary producers in marine/aquatic environments, where selenium is present, and in animals that are higher in the food web has led to the idea that other species—such as land plants—lost selenocysteine by necessity, as there is no or too little selenium available in their environment [14]. Given the huge importance of Ostreococcus tauri for carbon capture and marine food webs as a major species of marine phytoplankton, it is relevant to understand how their biochemistry is controlled by endogenous and environmental factors in the face of a changing environment. In addition to this direct relevance, Ostreococcus is now a bona fide model cell for the study of eukaryotic circadian rhythms that shares metabolic rhythms with human cells [22,29,38]. Therefore, this investigation can also directly inform studies on circadian regulation of the human or other vertebrate selenoproteomes, where a complete proteomic dataset would be harder to obtain.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cells11030340/s1: Figure S1: Protein abundance traces of all selenoproteins under LD and LL conditions. Figure S2: Amino acid usage among rhythmic versus arrhythmic proteins. Table S1: The Ostreococcus selenoproteome; proteome and transcriptome abundance profiles, protein properties, and rhythmicity parameters.
Author Contributions
Conceptualisation, G.v.O.; methodology, G.v.O. and H.K.; data analysis and figure preparation, H.K., H.T. and G.v.O.; experimentation, H.K. and G.v.O. manuscript preparation, H.K., H.T. and G.v.O. All authors have read and agreed to the published version of the manuscript.
Funding
H.K. and G.v.O. were supported by a Royal Society Research Fellows Enhancement Award to G.v.O. (RGF\EA\180192).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The mass spectrometry data were mined from publicly available data at the ProteomeXchange Consortium, with the dataset identifier PXD025009.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zhang E.E., Kay S.A. Clocks not winding down: Unravelling circadian networks. Nat. Rev. Mol. Cell Biol. 2010;11:764–776. doi: 10.1038/nrm2995. [DOI] [PubMed] [Google Scholar]
- 2.Covington M.F., Maloof J.N., Straume M., Kay S.A., Harmer S.L. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008;9:R130. doi: 10.1186/gb-2008-9-8-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang R., Lahens N.F., Ballance H.I., Hughes M.E., Hogenesch J.B. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc. Natl. Acad. Sci. USA. 2014;111:16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Putker M., Crosby P., Feeney K.A., Hoyle N.P., Costa A.S.H., Gaude E., Frezza C., O’Neill J.S. Mammalian Circadian Period, but Not Phase and Amplitude, Is Robust against Redox and Metabolic Perturbations. Antioxid. Redox. Signal. 2018;28:507–520. doi: 10.1089/ars.2016.6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Putker M., O’Neill J.S. Reciprocal Control of the Circadian Clock and Cellular Redox State—A Critical Appraisal. Mol. Cells. 2016;39:6–19. doi: 10.14348/molcells.2016.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lai A.G., Doherty C.J., Mueller-Roeber B., Kay S.A., Schippers J.H., Dijkwel P.P. CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proc. Natl. Acad. Sci. USA. 2012;109:17129–17134. doi: 10.1073/pnas.1209148109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edgar R.S., Green E.W., Zhao Y., van Ooijen G., Olmedo M., Qin X., Xu Y., Pan M., Valekunja U.K., Feeney K.A., et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;489:590. doi: 10.1038/nature11427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merrow M., Roenneberg T. Circadian clocks: Running on redox. Cell. 2001;106:141–143. doi: 10.1016/S0092-8674(01)00443-3. [DOI] [PubMed] [Google Scholar]
- 9.Spoel S.H., van Ooijen G. Circadian redox signaling in plant immunity and abiotic stress. Antioxid. Redox. Signal. 2014;20:3024–3039. doi: 10.1089/ars.2013.5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turner D.C., Stadtman T.C. Purification of protein components of the clostridial glycine reductase system and characterization of protein A as a selenoprotein. Arch. Biochem. Biophys. 1973;154:366–381. doi: 10.1016/0003-9861(73)90069-6. [DOI] [PubMed] [Google Scholar]
- 11.Arner E.S. Selenoproteins—What unique properties can arise with selenocysteine in place of cysteine? Exp. Cell Res. 2010;316:1296–1303. doi: 10.1016/j.yexcr.2010.02.032. [DOI] [PubMed] [Google Scholar]
- 12.Steinbrenner H., Speckmann B., Klotz L.O. Selenoproteins: Antioxidant selenoenzymes and beyond. Arch. Biochem. Biophys. 2016;595:113–119. doi: 10.1016/j.abb.2015.06.024. [DOI] [PubMed] [Google Scholar]
- 13.Stadtman T.C. Selenoproteins—Tracing the role of a trace element in protein function. PLoS Biol. 2005;3:e421. doi: 10.1371/journal.pbio.0030421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lobanov A.V., Hatfield D.L., Gladyshev V.N. Eukaryotic selenoproteins and selenoproteomes. Biochim. Biophys. Acta. 2009;1790:1424–1428. doi: 10.1016/j.bbagen.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palenik B., Grimwood J., Aerts A., Rouze P., Salamov A., Putnam N., Dupont C., Jorgensen R., Derelle E., Rombauts S., et al. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc. Natl. Acad. Sci. USA. 2007;104:7705–7710. doi: 10.1073/pnas.0611046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Derelle E., Ferraz C., Rombauts S., Rouze P., Worden A.Z., Robbens S., Partensky F., Degroeve S., Echeynie S., Cooke R., et al. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc. Natl. Acad. Sci. USA. 2006;103:11647–11652. doi: 10.1073/pnas.0604795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blanc-Mathieu R., Verhelst B., Derelle E., Rombauts S., Bouget F.Y., Carre I., Chateau A., Eyre-Walker A., Grimsley N., Moreau H., et al. An improved genome of the model marine alga Ostreococcus tauri unfolds by assessing Illumina de novo assemblies. BMC Genom. 2014;15:1103. doi: 10.1186/1471-2164-15-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corellou F., Schwartz C., Motta J.P., Djouani-Tahri el B., Sanchez F., Bouget F.Y. Clocks in the green lineage: Comparative functional analysis of the circadian architecture of the picoeukaryote ostreococcus. Plant Cell. 2009;21:3436–3449. doi: 10.1105/tpc.109.068825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Neill J.S., van Ooijen G., Dixon L.E., Troein C., Corellou F., Bouget F.-Y., Reddy A.B., Millar A.J. Circadian rhythms persist without transcription in a eukaryote. Nature. 2011;469:554–558. doi: 10.1038/nature09654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Ooijen G., Hindle M., Martin S.F., Barrios-Llerena M., Sanchez F., Bouget F.-Y., O’Neill J.S., Le Bihan T., Millar A.J. Functional Analysis of Casein Kinase 1 in a Minimal Circadian System. PLoS ONE. 2013;8:e70021. doi: 10.1371/journal.pone.0070021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Bihan T., Hindle M., Martin S.F., Barrios-Llerena M.E., Krahmer J., Kis K., Millar A.J., van Ooijen G. Label-free quantitative analysis of the casein kinase 2-responsive phosphoproteome of the marine minimal model species Ostreococcus tauri. Proteomics. 2015;15:4135–4144. doi: 10.1002/pmic.201500086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fustin J.M., Ye S., Rakers C., Kaneko K., Fukumoto K., Yamano M., Versteven M., Grunewald E., Cargill S.J., Tamai T.K., et al. Methylation deficiency disrupts biological rhythms from bacteria to humans. Commun. Biol. 2020;3:211. doi: 10.1038/s42003-020-0942-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kay H., Grunewald E., Feord H.K., Gil S., Peak-Chew S.Y., Stangherlin A., O’Neill J.S., van Ooijen G. Deep-coverage spatiotemporal proteome of the picoeukaryote Ostreococcus tauri reveals differential effects of environmental and endogenous 24-hour rhythms. Commun. Biol. 2021;4:1147. doi: 10.1038/s42003-021-02680-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sterck L., Billiau K., Abeel T., Rouze P., Van de Peer Y. ORCAE: Online resource for community annotation of eukaryotes. Nat. Methods. 2012;9:1041. doi: 10.1038/nmeth.2242. [DOI] [PubMed] [Google Scholar]
- 25.Boratyn G.M., Schaffer A.A., Agarwala R., Altschul S.F., Lipman D.J., Madden T.L. Domain enhanced lookup time accelerated BLAST. Biol. Direct. 2012;7:12. doi: 10.1186/1745-6150-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Binns D., Dimmer E., Huntley R., Barrell D., O’Donovan C., Apweiler R. QuickGO: A web-based tool for Gene Ontology searching. Bioinformatics. 2009;25:3045–3046. doi: 10.1093/bioinformatics/btp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Almagro Armenteros J.J., Salvatore M., Emanuelsson O., Winther O., von Heijne G., Elofsson A., Nielsen H. Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance. 2019;2 doi: 10.26508/lsa.201900429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zielinski T., Moore A.M., Troup E., Halliday K.J., Millar A.J. Strengths and limitations of period estimation methods for circadian data. PLoS ONE. 2014;9:e96462. doi: 10.1371/journal.pone.0096462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feeney K.A., Hansen L.L., Putker M., Olivares-Yanez C., Day J., Eades L.J., Larrondo L.F., Hoyle N.P., O’Neill J.S., van Ooijen G. Daily magnesium fluxes regulate cellular timekeeping and energy balance. Nature. 2016;532:375–379. doi: 10.1038/nature17407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robbens S., Derelle E., Ferraz C., Wuyts J., Moreau H., Van de Peer Y. The complete chloroplast and mitochondrial DNA sequence of Ostreococcus tauri: Organelle genomes of the smallest eukaryote are examples of compaction. Mol. Biol. Evol. 2007;24:956–968. doi: 10.1093/molbev/msm012. [DOI] [PubMed] [Google Scholar]
- 31.Hughes M.E., Hogenesch J.B., Kornacker K. JTK_CYCLE: An efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J. Biol. Rhythms. 2010;25:372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hutchison A.L., Maienschein-Cline M., Chiang A.H., Tabei S.M., Gudjonson H., Bahroos N., Allada R., Dinner A.R. Improved statistical methods enable greater sensitivity in rhythm detection for genome-wide data. PLoS Comput. Biol. 2015;11:e1004094. doi: 10.1371/journal.pcbi.1004094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novoselov S.V., Kryukov G.V., Xu X.M., Carlson B.A., Hatfield D.L., Gladyshev V.N. Selenoprotein H is a nucleolar thioredoxin-like protein with a unique expression pattern. J. Biol. Chem. 2007;282:11960–11968. doi: 10.1074/jbc.M701605200. [DOI] [PubMed] [Google Scholar]
- 34.Korotkov K.V., Kumaraswamy E., Zhou Y., Hatfield D.L., Gladyshev V.N. Association between the 15-kDa selenoprotein and UDP-glucose:glycoprotein glucosyltransferase in the endoplasmic reticulum of mammalian cells. J. Biol. Chem. 2001;276:15330–15336. doi: 10.1074/jbc.M009861200. [DOI] [PubMed] [Google Scholar]
- 35.Monnier A., Liverani S., Bouvet R., Jesson B., Smith J.Q., Mosser J., Corellou F., Bouget F.Y. Orchestrated transcription of biological processes in the marine picoeukaryote Ostreococcus exposed to light/dark cycles. BMC Genom. 2010;11:192. doi: 10.1186/1471-2164-11-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kojima S., Shingle D.L., Green C.B. Post-transcriptional control of circadian rhythms. J. Cell Sci. 2011;124:311–320. doi: 10.1242/jcs.065771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robles M.S., Cox J., Mann M. In-vivo quantitative proteomics reveals a key contribution of post-transcriptional mechanisms to the circadian regulation of liver metabolism. PLoS Genet. 2014;10:e1004047. doi: 10.1371/journal.pgen.1004047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Ooijen G., Millar A.J. Non-transcriptional oscillators in circadian timekeeping. Trends Biochem. Sci. 2012;37:484–492. doi: 10.1016/j.tibs.2012.07.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry data were mined from publicly available data at the ProteomeXchange Consortium, with the dataset identifier PXD025009.