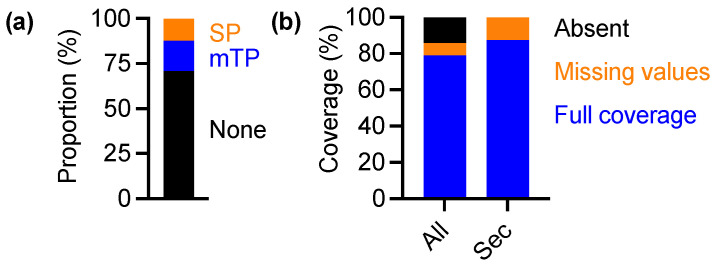
Figure 1.
Proteome coverage of the selenoproteome: (a) Proportion of selenoproteome predicted to have a transit peptide for the secretory pathway (SP), mitochondrion (mTP), or no transit peptide (None). (b) Coverage of proteins containing a selenocysteine (Sec) versus the whole proteome (All) in [23]. ‘Absent’ refers to proteins that were not quantified at any of the timepoints in either the LD or LL time series. ‘Missing values’ refers to proteins that were quantified in some but not all data points of the time series, while ‘Full coverage’ refers to those proteins that were reliably quantified at every timepoint throughout the LD and LL time series. We previously published detailed proteomic time series in Ostreococcus tauri under natural cycles of light and dark (LD; 12 h/12 h cycles of light/dark), as well as under constant light conditions (LL), sampling at 3.5 h intervals [23]. In that study, we obtained 85% coverage of the theoretical proteome and, interestingly, all 24 selenoproteins were detected in the dataset (Figure 1b and Supplementary Table S1). A total of 21 out of 24 selenoproteins had full coverage over the LD and LL time series (blue in Figure 1b), while 3 had missing data points (orange). Based on this exceptional coverage, we conclude that our dataset provides an unprecedented insight into rhythmic regulation of the selenoproteome.