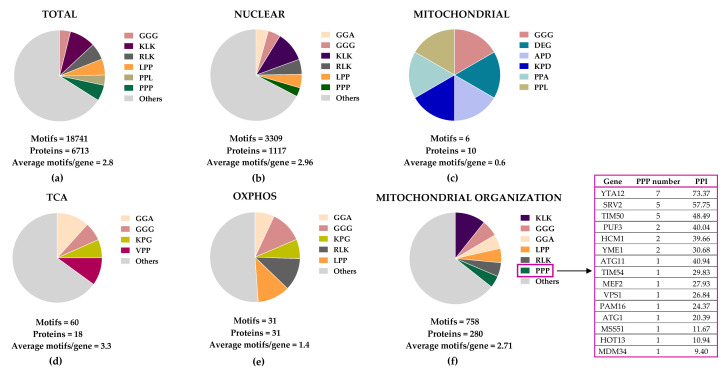
Figure 2.
Distribution of eIF5A-dependent motifs in Saccharomyces cerevisiae mitochondrial proteins. Distribution of the 43 highest-scoring eIF5A-dependent ribosome-pausing tri-peptide motifs [11] in the proteins of the whole yeast genome (a), nuclear-encoded mitochondrial proteins (b), mitochondrial-encoded proteins (c), the tricarboxylic acid (TCA) cycle (d), oxidative phosphorylation (OXPHOS) (e), and in the mitochondrial organization Gene Ontology functional category (f). The table shows the proteins involved in mitochondrial organization with at least one PPP motif. The protein pause index (PPI) is calculated as the sum of the quantitative value of the ribosome pause provoked by depletion of eIF5A in each of the 43 highest eIF5A-dependent tri-peptide motifs [11] found in the amino acid sequence of each protein, and is higher in proteins that are putatively more dependent on eIF5A for their translation.