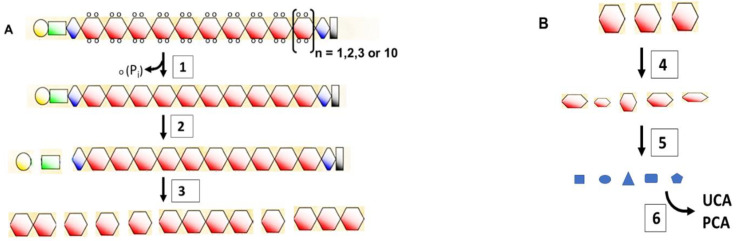
Figure 4.
Flowchart of the processing of profilaggrin to free amino acids. (A). Proflaggrin to filaggrin. (1) Phospho-profilaggrin is dephosphorylated by phosphatases. (2) The A (yellow) and B (green) domains are cleaved from profilaggrin by furin, PACE4 (Paired basic Amino acid Cleaving Enzyme 4) and endoproteinase-1 (PEP1) [50,51]. (3) Linker sequences of human profilaggrin (FLYQVST) are cleaved by skin-specific retrovirus-like aspartic protease (SASPase) [52], channel-activating serine protease (CAP1) [53] and matriptase (MT-SP1) [54] to monomeric filaggrin. Aminopeptidases and carboxypeptidases are likely involved in trimming the new termini [37]. (B). Filaggrin to NMF. (4) Arginine residues in filaggrin and keratin are converted to citrulline by peptidylarginine deiminase (PAD1 or 3) [49,55]. Deiminated filaggrin is cleaved by calpain-1 and caspase-14 (at VSQD and HSED sequences) to filaggrin fragments [55,56,57]. (5) Filaggrin fragments are digested by neutral cysteine protease (bleomycin hydrolase) to amino acids [57]. Aminopeptidases and carboxypeptidases are also likely involved. (6) Histidine is converted by histidine deaminase to trans-urocanic acid (UCA), which has a UV spectrum similar to that of nucleic acids and proteins [58] and provides a natural sunscreen [59]. Glutamine is non-enzymatically converted to 2-pyrrolidone carboxylic acid (PCA). These hydrophilic final products contribute to the moisturizing of the skin [56,57,60,61]. Kezic et al. [62] and de Veer et al. [63] provided excellent reviews of proteolytic processing of filaggrin and differentiation of corneocytes.