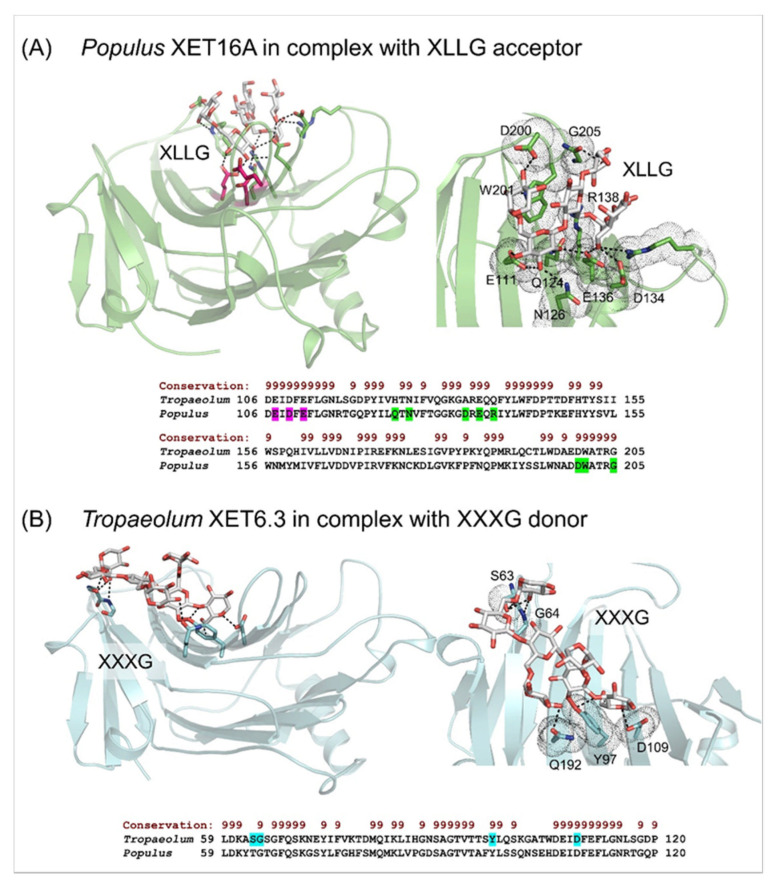
Figure 2.
β-Sandwich architectures and β-jelly-roll topologies of Populus XET16A (PDB accession 1UMZ) in complex with the XLLG acceptor substrate (A), and the Tropaeolum XET6.3 3D molecular model in complex with the XXXG donor substrate (B). (A) Left panel: The XLLG acceptor (cpk sticks) bound in the Populus XET16A structure is indicated in dashed lines at 2.6 Å to 3.5 Å separations. The catalytic trio (Glu85, Glu89 and Asp87) is shown in cpk magenta sticks. Right panel: Details of the XLLG binding in Populus XET16A; interacting residues are marked in green cpk sticks and dots. Bottom panel: Some of the residues of Populues XET16A binding XLLG (highlighted in green) are shown in the alignment by PROMALS3D [83] of the Populus and Tropaeolum XET sequences (numbering includes signal peptides). Conservation of residues on the scale 9–6 is shown on the top of the alignment in brown. (B) Left panel: The XXXG donor (cpk sticks) bound in Tropaeolum XET6.3 (cyan; coordinates from [66]) is indicated in dashed lines at separations between 2.3 Å and 3.0 Å. Right panel: Details of XXXG binding in Tropaeolum XET6.3; interacting residues are marked in cyan cpk sticks in dots. Bottom panel: Some of the residues of Tropaeolum XET6.3 binding XLLG (highlighted in cyan) are shown in the alignment by PROMALS3D [83] of Populus XET16A and Tropaeolum XET6.3 sequences (numbering includes signal peptides). Conservation of residues on the scale 9–6 is shown on the top of the alignment in brown. Images were generated in the PyMOL Molecular Graphics System v2.5.2 (Schrődinger LLC, Portland, OR, USA).