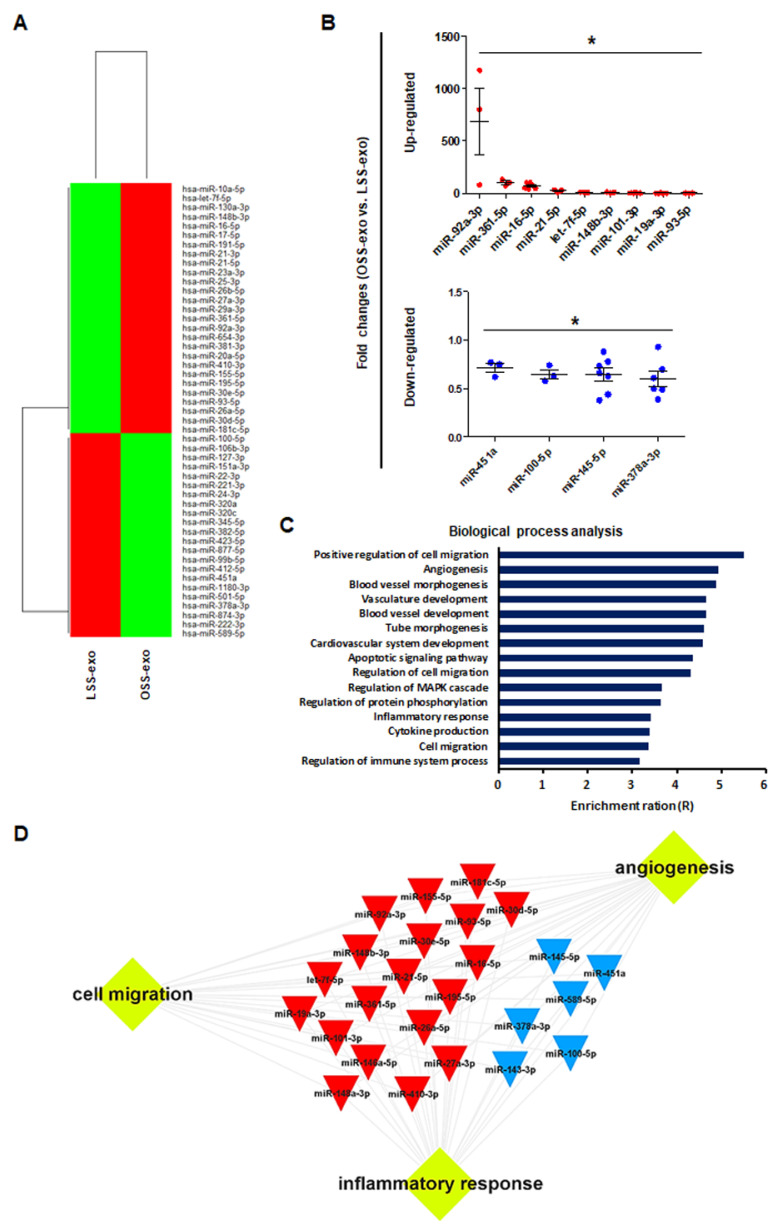
Figure 2.
Biological processes targeted by the miRNA landscape in shear stress-induced EC-sEVs. (A) Heat map showing upregulated and downregulated miRNAs in shear stress-induced EC-sEVs by miRNA sequencing. A total of 39 upregulated miRNAs and 43 downregulated miRNAs in OSS-sEVs compared with LSS-sEVs were identified (LSS-sEVs or OSS-sEVs; n = 3). (B) A total of 82 miRNAs selected from miRNA sequencing analysis were validated by qRT-PCR. A total of 13 miRNAs were significantly differentially expressed between OSS-sEVs and LSS-sEVs: 9 upregulated (upper) and 4 downregulated (lower) miRNAs (n = 4; compared to LSS-sEVs, * p < 0.05). Mean ± SEM. Statistical analysis was performed using the nonparametric Mann–Whitney U test. (C) To identify enriched biological processes targeted by the validated 13 miRNAs in shear stress-induced EC-sEVs, miRTarBase 7.0 was used as the basis for miRNA–mRNA interactions. The miRNA target gene set was analyzed using the WEB-based Gene SeT AnaLysis Toolkit (WebGestalt) for biological process analysis. (D) A typical 13 miRNAs-target network visualized by WebGestalt.