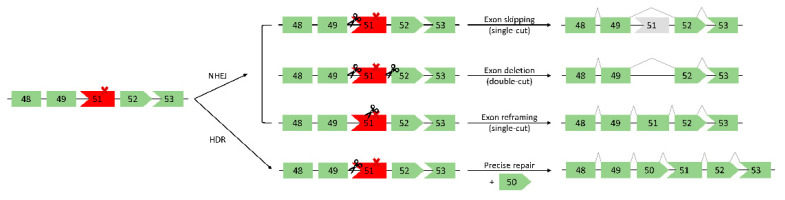
Figure 3.
A schematic of DNA repair systems for gene editing with CRISPR/Cas9 in a hypothetical DMD patient harboring an exon 50 deletion mutation. Deleting exon 50 abolishes the reading frame and leads to a premature stop codon in exon 51, denoted by the “X”. Non-homologous end joining (NHEJ) or homology-directed repair (HDR) can restore the dystrophin reading frame. NHEJ exon skipping (single-cut) targets a splice site, thus skipping the adjacent exon. NHEJ exon deletion (double-cut) requires two sgRNAs flanking the out-of-frame exon(s) for removal. Exon reframing (single-cut) relies on indels generated by NHEJ to reframe the out-of-frame exon. Finally, HDR requires an exogenous DNA template to replace the missing or mutated exon in a precise manner. Scissors represent the sites targeted by CRISPR/Cas9.