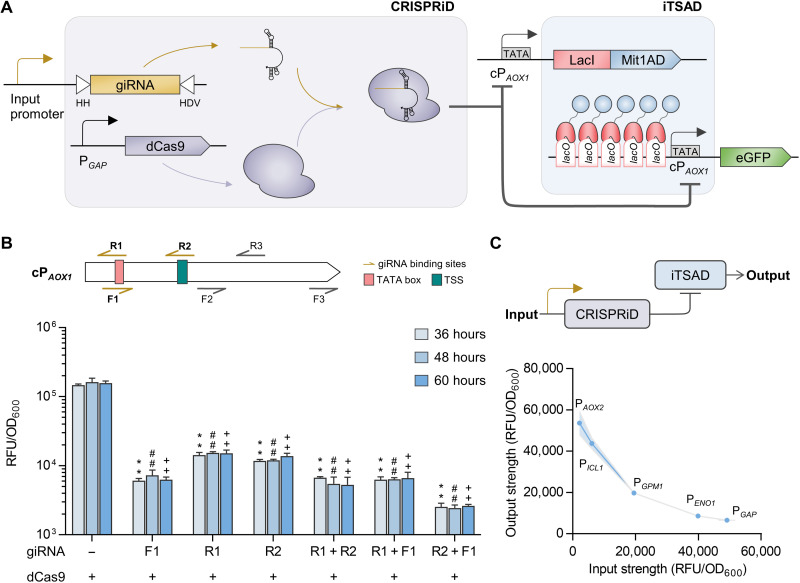
Fig. 2. Development of CRISPRiD to suppress the leaky expression of iTSAD.
(A) Schematic diagram illustrating repression of CRISPRiD on iTSAD. The giRNA was designed to target cPAOX1, which drives both iTSAD and eGFP, thereby recruiting dCas9 to achieve dual suppression of iTSAD by CRISPRiD. (B) Repressive effect of CRISPRiD with different giRNAs on iTSAD. Six giRNAs targeting various sites with a 5′-NGG-3′ PAM sequence across the cPAOX1 were designed to suppress cPAOX1 activity. Detailed information of the binding sites and repressive effects of the giRNAs is elaborated in fig. S2. The three best giRNAs (giRNA_F1, giRNA_R1, and giRNA_R2) and their pairwise combinations were selected to suppress the leaky expression of iTSAD. Statistical significance of eGFP expression in strains with various giRNAs relative to the parent strain without a giRNA at each time point is shown (**P < 0.01 at 36 hours; ##P < 0.01 at 48 hours; ++P < 0.01 at 60 hours). (C) The relationship of input strength and output strength of the CRISPRiD-iTSAD tandem system. A simplified diagram of CRISPRiD-iTSAD system is shown. Five promoters with various strengths were set as input promoters to test the repressive function of CRISPRiD on iTSAD. The regression model is shown in fig. S2. RFU, relative fluorescence unit.