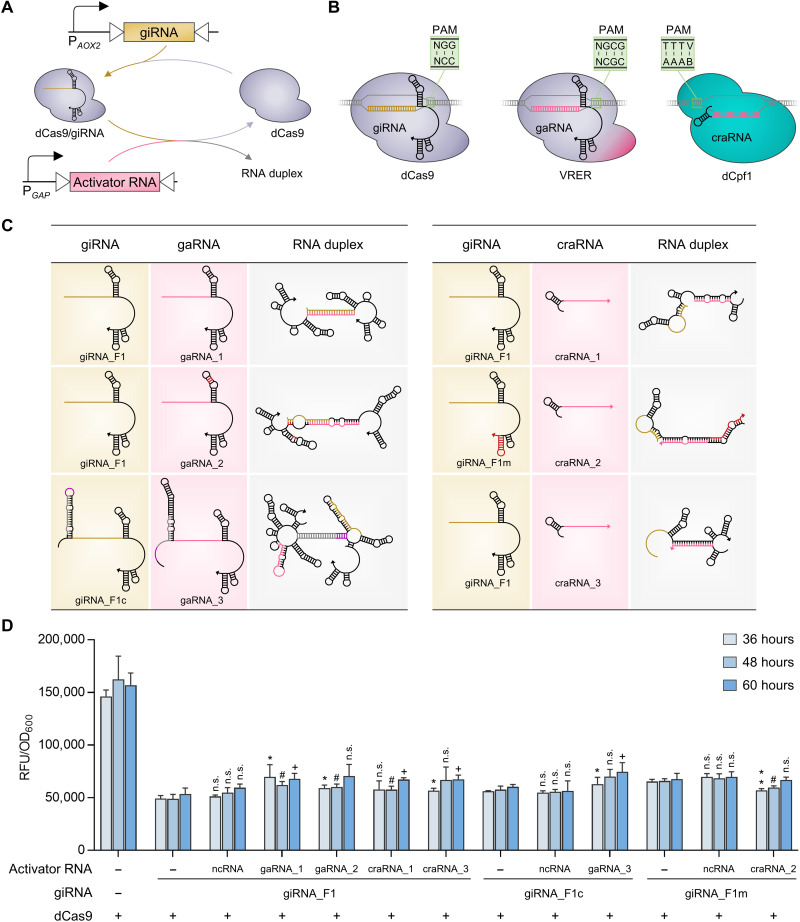
Fig. 3. Design of activator RNAs and their derepression effects on CRISPRiD.
(A) Schematic diagram of activator RNA–mediated interference of CRISPRiD repression. The activator RNA and giRNA were driven by PGAP and PAOX2, respectively. PGAP is much stronger than the PAOX2. (B) dCas regulators orthogonal to dCas9, which recognizes 5′-NGG-3′ PAM sequences, were selected, which were adapted to activator RNAs. These dCas regulators were selected keeping the design of CRISPRa, which is depicted in Fig. 4, in mind. Two dCas regulators were selected to avoid cross-talk between CRISPRa and CRISPRi. The S. pyogenes dCas9 mutant VRER (D1135V/G1218R/R1335E/T1337R), which recognizes 5′-NGCG-3′ PAM sequences, and L. bacterium–derived dCpf1 (E832A), which recognizes 5′-TTTV-3′ PAM sequences, were used. (C) Design of activator RNAs and secondary structure predictions for the duplex giRNAs and activator RNAs (left, gaRNA; right, craRNA). Three gaRNAs and three craRNAs were designed to dimerize with the corresponding giRNA, thereby interfering with the repression of CRISPRiD. The arrow points to the 3′ end of the RNA. Details of RNA design and interactions are described in the Supplementary Materials. (D) Derepression effects on CRISPRiD by the interaction of activator RNA with giRNA. The ncRNA refers to a short RNA without an interaction region with giRNA_F1, giRNA_F1c, or giRNA_F1m. Statistical significance of eGFP expression of each strain with activator RNA relative to the parent strain without activator RNA is shown for each time point (**P < 0.01 and *P < 0.05 at 36 hours; #P < 0.05 at 48 hours; +P < 0.05 at 60 hours; n.s., not significant). RFU, relative fluorescence unit.