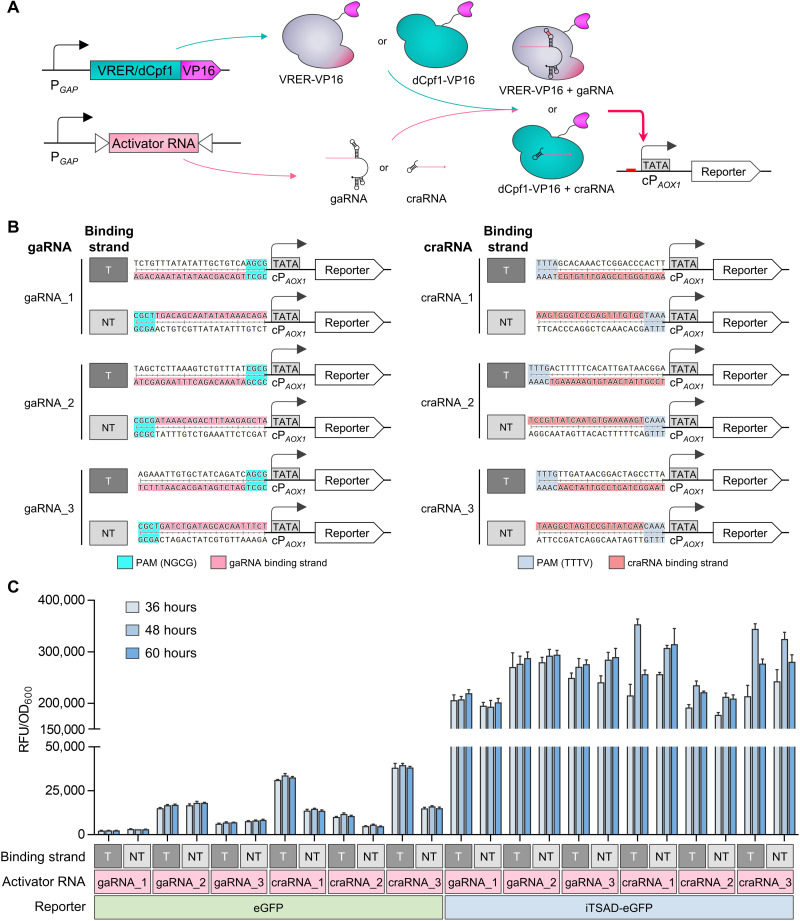
Fig. 4. Design and characterization of CRISPRaD based on VRER or dCpf1.
(A) Schematic diagram illustrating the activation effect of CRISPRaD on cPAOX1-driven reporters. The binding sequences of gaRNAs or craRNAs were inserted upstream of cPAOX1. The viral activator VP16 was fused to the C terminus of VRER or dCpf1, and it recruited RNA polymerase. The reporters are eGFP or iTSAD-driven eGFP. (B) Design of various sequences inserted upstream of the cPAOX1 for the recognition and binding of activator RNA (gaRNA and craRNA). T represents activator RNA binding to the transcribed strand, and NT represents activator RNA binding to the nontranscribed strand. The sequences of gaRNA or craRNA binding sites are highlighted in pink and red, respectively. The PAM sequences of gaRNA and craRNA are highlighted in cyan and blue, respectively. (C) Activation effects of CRISPRaDs on the cPAOX1-driven reporters with different activator RNAs. Each activator RNA was designed to bind with the T-strand and NT-strand by targeting the same base-pairing region. The fluorescence intensities of eGFP and iTSAD-driven eGFP as reporters are shown. RFU, relative fluorescence unit.