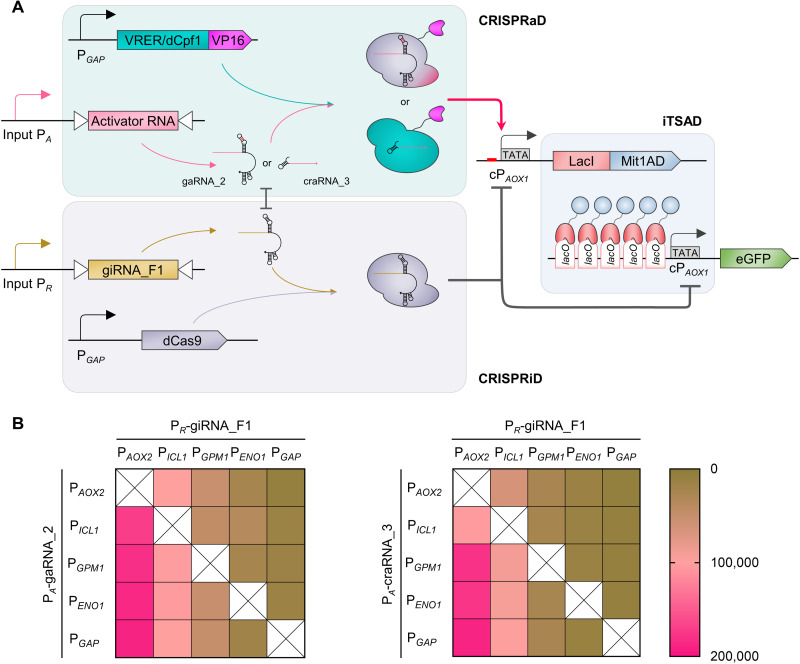
Fig. 5. Characterization of a synthetic transcription regulatory system integrating iTSAD, CRISPRiD, and CRISPRaD.
(A) Schematic diagram illustrating the assembly of iTSAD, CRISPRiD, and CRISPRaD. Two kinds of CRISPRaD, mediated by VRER-VP16/gaRNA_2 and dCpf1-VP16/craRNA_3, were tested. For this integrated system, the cPAOX1-driven iTSAD was activated by PA-driven CRISPRaD and repressed by PR-driven CRISPRiD. The PA and PR can be selected to respond to various signals, so they can be user-defined. As shown in Fig. 4B, gaRNA_2 was designed to bind the NT-strand upstream of cPAOX1, and craRNA_3 was designed to bind the T-strand upstream of cPAOX1. Activator RNA (gaRNA_2 or craRNA_3) of CRISPRaD mildly interfered with giRNA_F1 of CRISPRiD as shown in Fig. 3D. (B) Heatmaps showing the output strength of the synthetic transcription regulatory system with various input strengths of PA and PR. Left, system with CRISPRaD based on VRER-VP16/gaRNA_2; right, system with CRISPRaD based on dCpf1-VP16/craRNA_3. Five promoters with increasing native strength—that is, PAOX2, PICL1, PGPM1, PENO1, and PGAP—were used as PA and PR. A total of 20 pairwise combinations of these promoters for PA and PR were designed, and 40 strains with different synthetic transcription regulatory systems were constructed. The eGFP expression of the different strains was measured after being cultured for 48 hours in YPD medium.