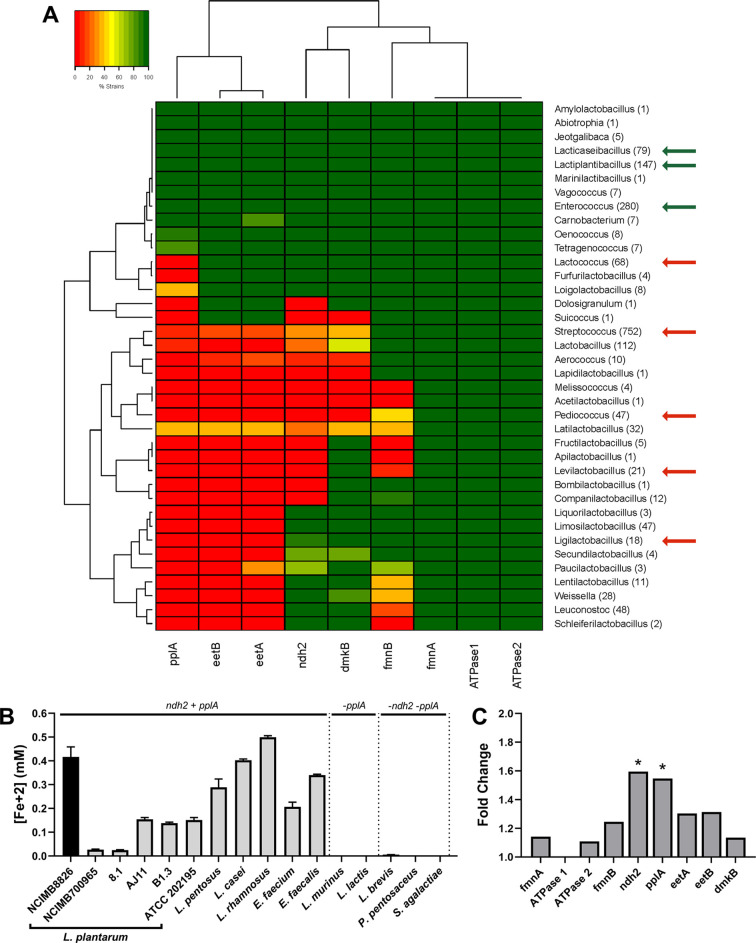
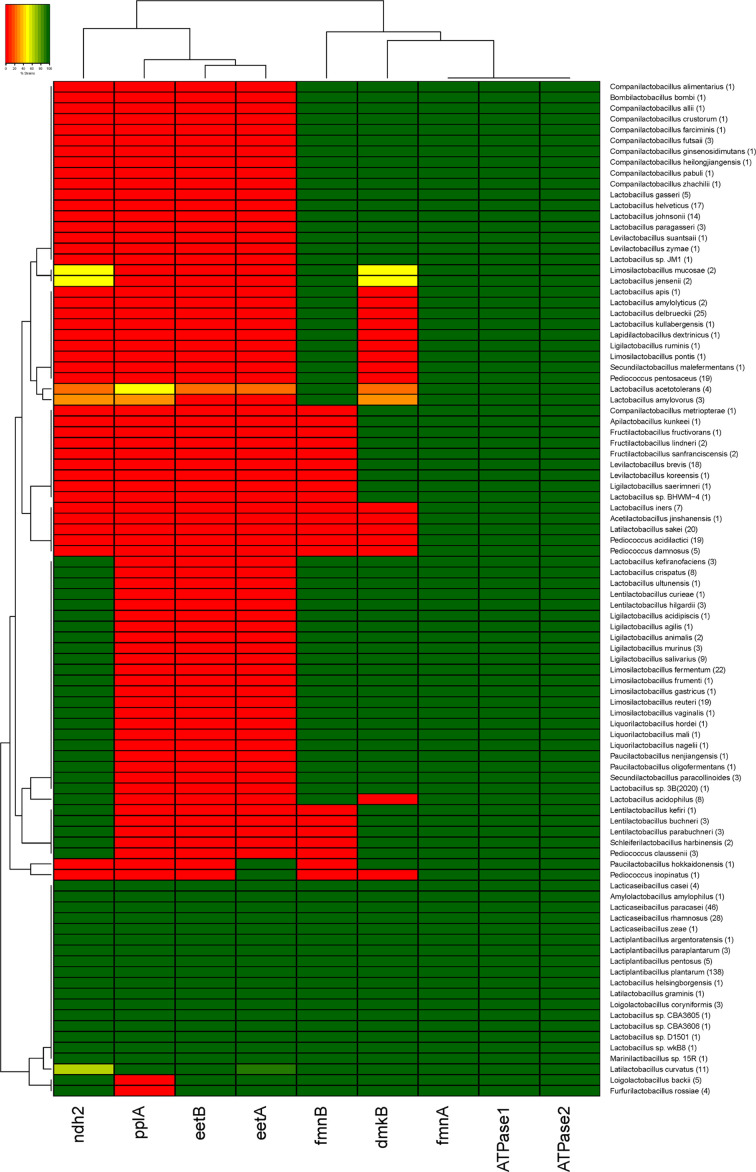
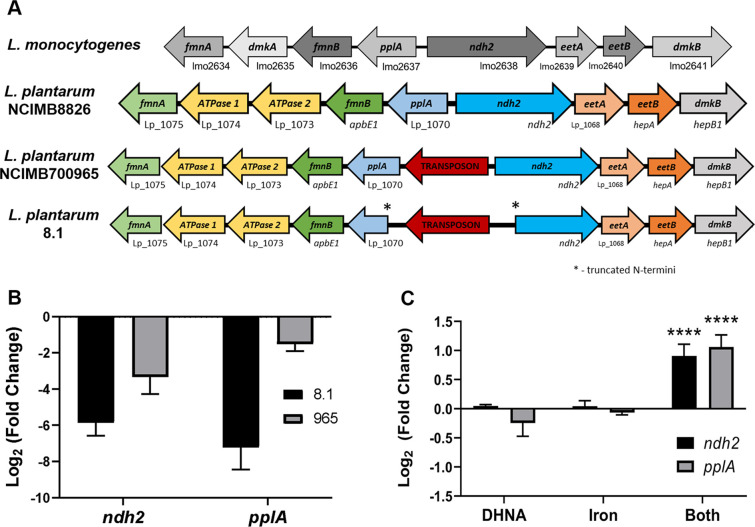
Figure 2. The FLEET genes ndh2 and pplA are associated with iron reduction by LAB.
(A) Heatmap showing the genera in the Lactobacillales order containing FLEET genes. Homology searches were conducted using tBLASTx for 1788 complete LAB genomes in NCBI (downloaded 02/25/2021) against the L. plantarum NCIMB8826 FLEET locus. A match was considered positive with a Bit-score >50 and an E-value of <10–3. Arrows designate genera tested for iron reduction activity; green = EET-active with Fe3+, red = EET-inactive with Fe3+. (B) Reduction of ferrihydrite in PBS with 20 μg/mL DHNA and 55 mM mannitol after growth in mMRS supplemented with 20 μg/mL DHNA and 1.25 mM ferric ammonium citrate. The avg ± stdev of three biological replicates per strain is shown. (C) Relative expression of NCIMB8826 FLEET locus genes in mMRS with 20 μg/mL DHNA and 1.25 mM ferric ammonium citrate compared to growth in mMRS. Significant differences in expression were determined by the Wald test (n = 3) with a Log2 (fold change) > 0.5 and an FDR-adjusted p-value of <0.05. See also Figure 2—figure supplement 1 and Figure 2—figure supplement 2 and related data in Figure 2—source data 1.