Figure 1.
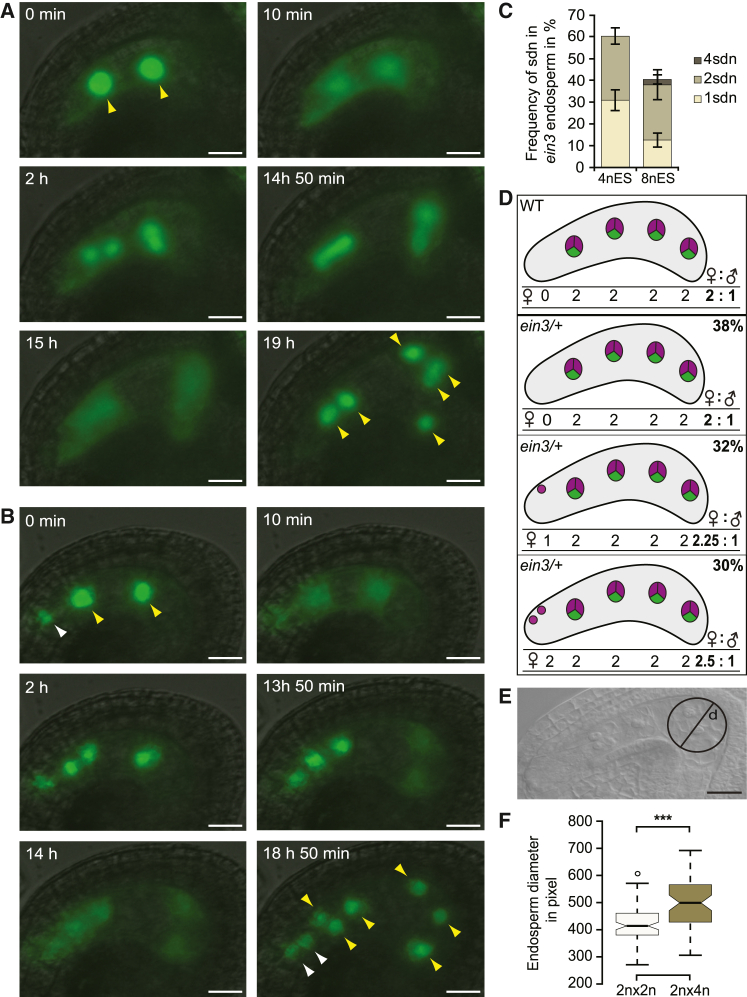
Ploidy differences in the seed can be detected at the zygote stage.
(A and B) Live-cell imaging of young ein3/− × wild-type seeds without (A) or with (B) sdn. The combinatorial multicolor marker FGR 7.0 confers green fluorescence to sdn and endosperm nuclei. Endosperm nuclei, yellow arrowheads; sdn, white arrowheads. See also Supplemental Videos 1 and 2.
(C) Frequency of one, two, or four sdn in ein3/− × wild-type seeds at the four- (4nES) and eight-nucleate endosperm stage (8nES) (n = 136/166 for 4nES/8nES respectively).
(D) Schematic representation indicating the changes in maternal- (magenta) to-paternal (green) genome ratio in the four-nucleate endosperm of a plant recovered from an ein3/- x wild-type cross containing no sdn, one sdn, or two sdn (n = 338).
(E) Wild-type seed in the four-nucleate endosperm stage. Dorsoventral endosperm diameter was determined by superimposing a circle inside the endosperm at its widest point using ImageJ.
(F) Dorsoventral endosperm diameter of interploidy crosses in wild-type seeds at the four-nucleate endosperm stage (n (2n × 2n) = 84; n (2n × 4n) = 58). Data are the mean ± SEM. Scale bar, 20 μm. Two-tailed Student’s t-test: ∗∗∗p < 0.001.