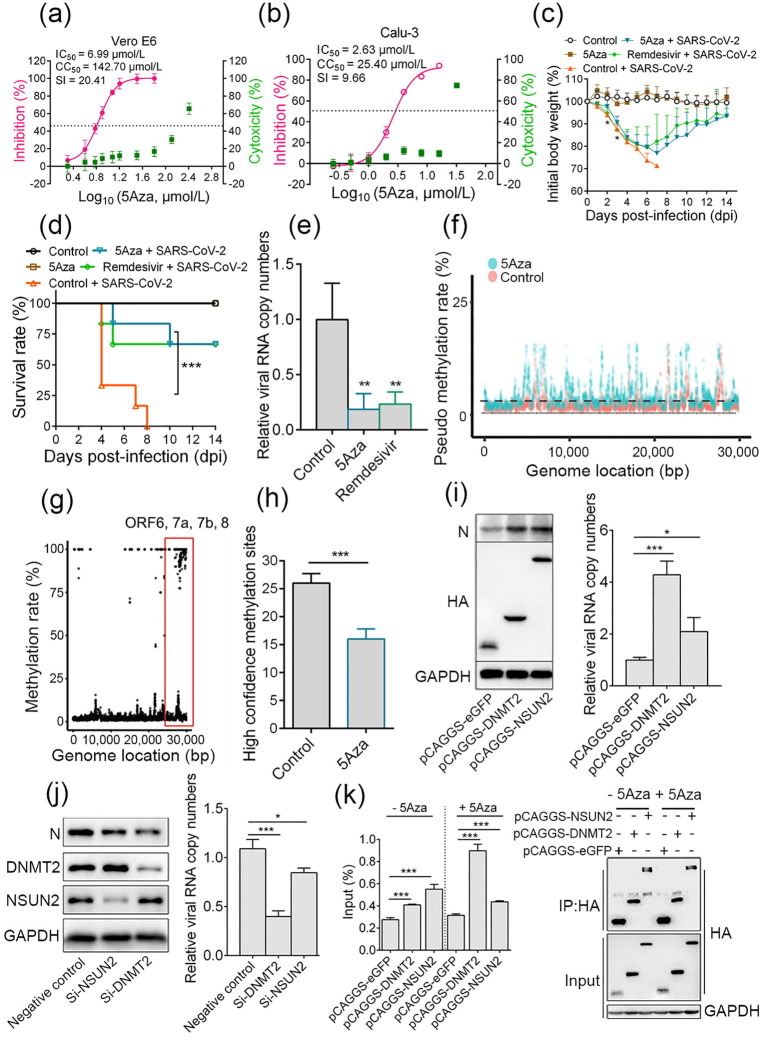
Fig. 1.
Azacytidine targets SARS-CoV-2 RNA to inhibit virus infection. Vero E6 and Calu-3 cells were infected with SARS-CoV-2 at a multiplicity of infection (moi) of 0.2 in the presence of different doses of azacytidine. At 24 h post-infection, cell supernatants were collected. (a, b) IC50 and CC50 were calculated by detecting viral RNA with quantitative real-time PCR (qRT-PCR) and CCK-8 assay, respectively. (c–e) The anti-SARS-CoV-2 effect of azacytidine in vivo. 8 weeks old female BALB/c mice were randomly divided into 3 groups with 10 mice per group. Mice were intranasally challenged with 4 × 103 plaque forming unit (PFU) MA-SARS-CoV-2 in 50 μL Dulbecco’s modified Eagle’s medium (DMEM) or equal DMEM. At 1 dpi, mice were intraperitoneally injected with either 2 mg/kg 5Aza, 25 mg/kg Remdesivir, or an equivalent volume of sterile saline, once daily for seven consecutive doses. (c) Body weight was measured daily for 14 d; mice with 30% body weight loss were considered moribund and euthanized. (d) The survival rates of mice (n = 6). (e) At 4 dpi, 4 mice per group were euthanized to detect viral RNA copy numbers in lungs, and data are presented as values relative to the control. (f) The pseudo m5C locations indicated incorporated azacytidine. Vero E6 cells were infected with 1 moi SARS-CoV-2 in the presence of 10 μmol/L 5Aza or saline. After 12 h, total RNA was isolated and a comparison of 5Aza-treated viral RNA with the control using bisulfite sequencing was performed; the non-overlapping points are pseudo m5C locations that indicate where 5Aza was incorporated. (g) RNA-BSseq identified m5C locations in the SARS-CoV-2 genome. There were at least 5 methylation-supporting reads per point. Data are averaged from 3 technological repeats. (h) RNAs of cells infected with SARS-CoV-2 (treated with 5Aza or untreated) were subjected to nanopore direct RNA sequencing. (i) Vero E6 cells were transiently overexpressed with DNMT2, NSUN2, or GFP; 24 h later, cells were infected with 0.1 moi SARS-CoV-2. After 24 h, the supernatant was collected for viral RNA detection by qRT-PCR, and cells were lysed and subjected to western blotting. (j) Vero E6 cells were transfected with 100 μmol/L siRNA targeting NSUN2, DNMT2, or negative control siRNA; 48 h later, cells were infected with 0.1 moi SARS-CoV-2. After 30 h, viral RNA in the supernatant was detected by RT-PCR, and cells were lysed for western blotting. (k) Vero E6 cells transiently overexpressing DNMT2, NSUN2, or GFP with a hemagglutinin (HA) tag were infected with 0.2 moi SARS-CoV-2 for 20 h, either in the presence of 16 μmol/L 5Aza or not. Lysates were prepared and split for incubation with mouse anti-HA antibody. Co-precipitated RNA was analyzed by qRT-PCR. The level of viral RNA amplicon was determined as the percentage of input (1% of lysate) (left); the expression of indicated protein and products of immunoprecipitation (IP) was validated by western blotting (right). Data are presented as mean ± standard deviation and analyzed using Student’s t-test or one-way analysis of variance (body weight change); Log-rank test was used to analyze the significance of survival differences. * P < 0.05, ** P < 0.01, *** P < 0.001.