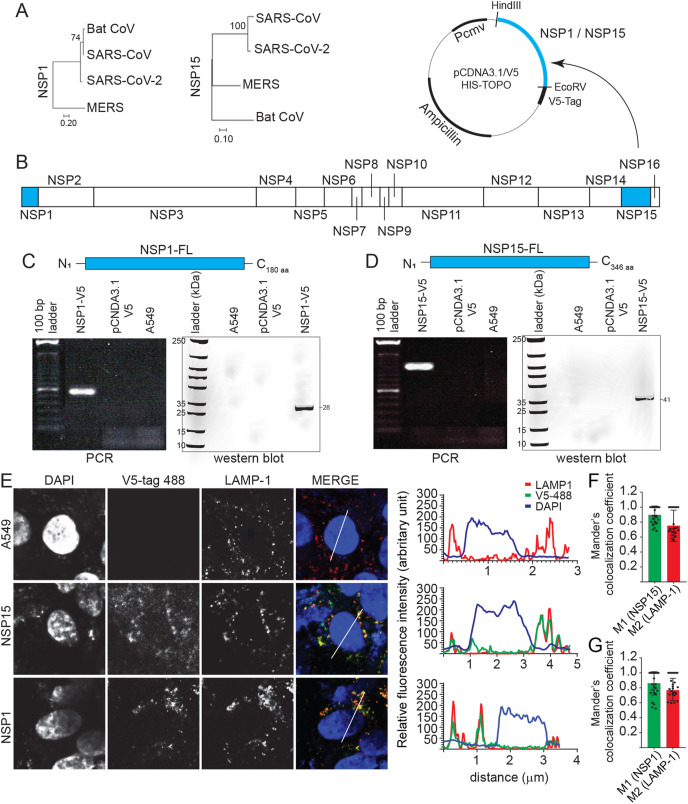
Fig. 1.
NSP1 and NSP15 stably expressing A549 cells lines. Phylogenetic analysis of the SARS-CoV-2 (A) NSP1 and NSP15 as performed using MEGAX software. (B) NSP1 and NSP15, part of the ORF1a and 1b were subcloned into a pCDNA3.1/V5–HIS-TOPO plasmid (HindIII and EcoRV restriction sites) and transfected into A549 cells. Expression of (C) NSP1 and (D) NSP15 was confirmed by total RNA extraction (polymerase chain reaction; PCR) and total protein extraction (Western blot). (E) Representative pictures of confocal microscopy imaging with maximum projections of z-stacks for each channel demonstrating the subcellular localization of expressed NSP1 (anti-V5 antibody; 488 nm) and NSP15 (anti-V5 antibody; 488 nm) and lysosome-associated membrane protein 1 (LAMP-1) (red) in stably expressing A549 cells. Maximum projections of all combined channels (merge) were analyzed using LAS AF software to determine the relative fluorescence intensity (arbitrary units [A.U]) profiles in stably expressing cell line for anti-V5 (green) and DAPI (blue) and LAMP-1 (red) along the white line path (micrometers) indicated in the corresponding ROIs (merge) demonstrating overlapping signals. Validation of colocalization efficiencies by Manders’ colocalization coefficient as analyzed using ImageJ 72 h. Colocalizations between (F) NSP1-anti-V5 (M1) and LAMP-1 (M2) and (G) NSP15-anti-V5 (M1) and LAMP-1 (M2) represent the overlap of M1 as the denominator and vice versa (n = 50). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)