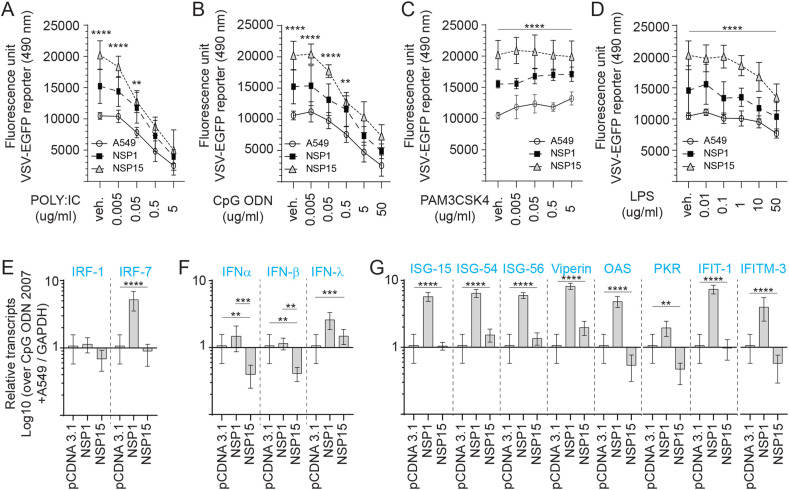
Fig. 4.
TLR ligand sensitivity of NSP1 and NSP15 stably expressing A549 cells. Replication efficiency of the interferon sensitive VSV-EGFP virus in A549 cells. Analysis of VSV-EGFP (moi = 0.4) virus replication in NSP1 and NSP15 stably expressing A549 cells lines compared to control transfected (pCDNA3.1/V5–HIS-TOPO) or negative (A549 cells) based on fluorescence (EGFP; excitation 490 nm). Cell lines were pre-stimulated (2 h) with (A) TLR3 ligand POLY; IC (0.005, 0.05, 0.5 and 5 μg/mL), (B) TLR9 ligand CpG ODN 2007 (0.005, 0.05, 0.5 and 5 μg/mL), (C) TLR2 ligand PAM3CSK4 (0.005, 0.05, 0.5 and 5 μg/mL), and (D) TLR4 ligand LPS (0.01, 0.1, 1.0, 10 and 50 μg/mL) prior to infection with VSV-EGFP. Fold changes in (E) IRF1 and IRF7, (F) PAN-IFN-α, IFN-β and IFN-γ, and (G) ISG-15, ISG-54, ISG-56, Viperin, OAS, PKR, IFIT1 and IFITM-3 gene expression based on qRT-PCR in NSP1 and NSP15 stably expressing A549 cells lines over negative control (A549 cells) stimulated for 2 h with 5 μg/mL CpG ODN 2007. The mean fluorescence unit in uninfected cells was used as the normalization factor for VSV-EGFP readout in infected A549 cells.Non-parametric Wilcoxon tests (Mann-Whitney) and one-way ANOVA was used to assess normal distribution and test significance with the results shown as mean ± SD. ∗∗ (p ≤ 0.01), ∗∗∗ (p ≤ 0.005) and ∗∗∗∗ (p ≤ 0.001) indicates a statistically significant difference. All viral infection experiments were performed in 10 technical replicates, and the data are representative of results from 3 independent experiments.