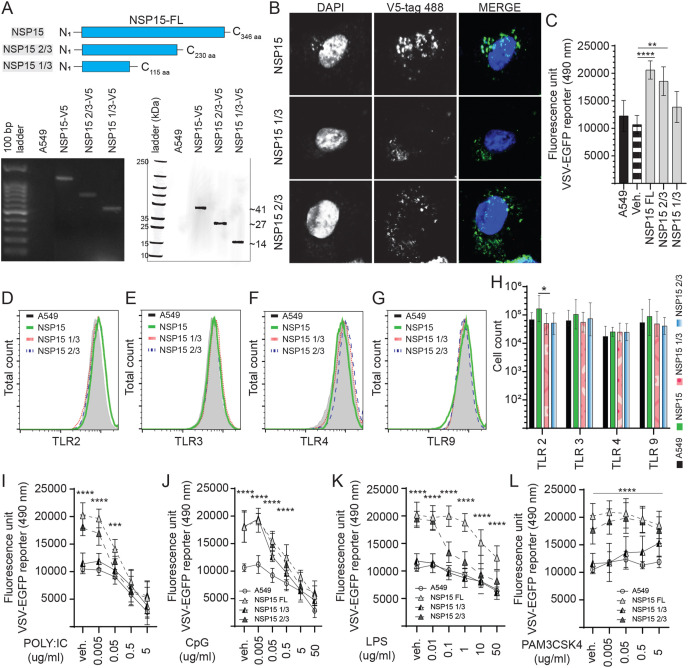
Fig. 6.
Identification of the NSP15 protein functional domain. Truncation of NSP15 limits VSV-EGFP virus replication. (A) Schematic diagram depicting the full length of NSP15 and truncated NSP15 1/3 and NSP15 2/3 in amino acid. The SARS-CoV-2 NSP15 was truncated by high fidelity PCR (GoTaq) and subcloned into a pCDNA3.1/V5–HIS-TOPO plasmid (HindIII and EcoRV insertion sites) which was transfected into A549 cells to generate stably expressing cell lines. Expression were confirmed by total RNA extraction (polymerase chain reaction; PCR) and total protein extraction (Western blot). (B) Representative pictures of confocal microscopy imaging with maximum projections of z-stacks for each channel demonstrating the subcellular localization of expressed full length of NSP15, NSP15 1/3 and NSP15 2/3 (anti-V5 antibody; 488 nm) in stably expressing A549 cells. Analysis of VSV-EGFP (moi = 0.4) virus replication in (C) NSP15, NSP15 1/3 and NSP15 2/3 stably expressing A549 cells lines compared to control transfected (pCDNA3.1/V5–HIS-TOPO) or negative (A549 cells) based on fluorescence (EGFP; excitation 490 nm). Representative overlapping FACS histogram demonstrating the expression of (D) TLR2 (E) TLR3, (F) TLR4 and (G) TLR9 as gated on live (7AAD-) NSP15, NSP15 1/3 and NSP15 2/3 stably expressing A549 cells lines cells to analyze (H) the number of cells expressing TLR2, TLR3, TLR4 and TLR9 compared to negative control (A549 cells). Analysis of VSV-EGFP (moi = 0.4) virus replication in NSP15, NSP15 1/3 and NSP15 2/3 stably expressing A549 cells lines compared to control transfected (pCDNA3.1/V5–HIS-TOPO) or negative (A549 cells) based on fluorescence (EGFP; excitation 490 nm) pre-stimulated (2 h) with (I) TLR3 ligand polyI:C (0.005, 0.05, 0.5 and 5 μg/mL), (J) TLR9 ligand CpG ODN 2007 (0.005, 0.05, 0.5 and 5 μg/mL), (K) TLR4 ligand LPS (0.01, 0.1, 1.0, 10 and 50 μg/mL) and (L) TLR2 ligand PAM3CSK4 (0.005, 0.05, 0.5 and 5 μg/mL) and prior to infection with VSV-EGFP. The mean fluorescence unit in uninfected cells was used as the normalization factor for VSV-EGFP readout in infected A549 cells. Non-parametric Wilcoxon tests (Mann-Whitney) or one-way ANOVA was used to assess normal distribution and test significance with the results shown as mean ± SD. ∗ (p ≤ 0.05), ∗∗ (p ≤ 0.01), ∗∗∗ (p ≤ 0.005) and ∗∗∗∗ (p ≤ 0.001) indicates a statistically significant difference. All viral infection experiments were performed in 10 technical replicates, and the data are representative of results from 3 independent experiments.