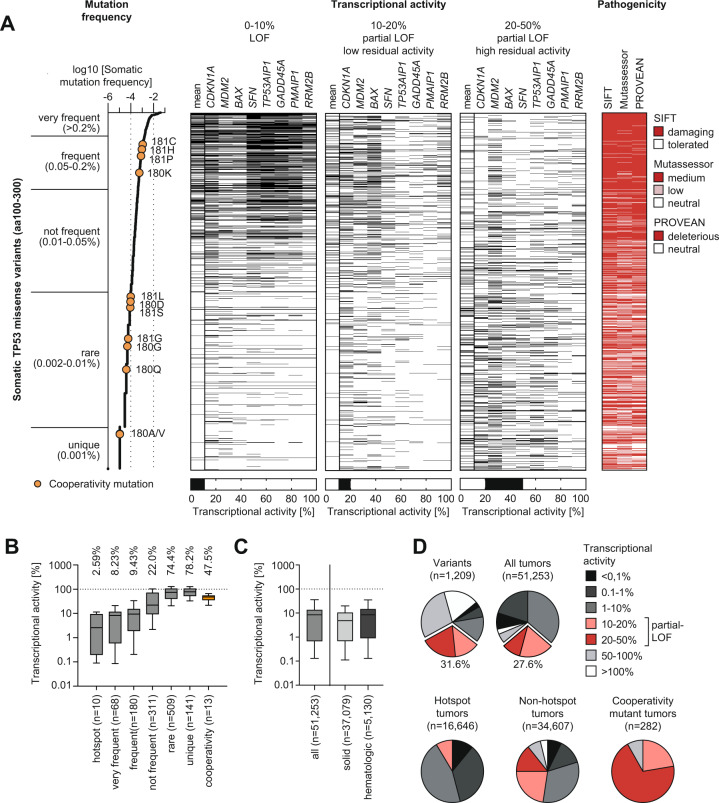
Fig. 1. Prevalence of p53 partial-LOF mutations in human cancer patients.
A Frequency, transcriptional activity, and in silico predicted pathogenicity of 1,209 somatic TP53 missense mutations affecting the p53 DNA binding domain (amino acids 100–300). Three black-white heatmaps depict the mean and response element-specific transcriptional activity of missense variants. The left heatmap illustrates the distribution of loss-of-function (LOF) events (0-10% transcriptional activity), the middle heatmap partial LOF events with low (10–20%) residual activity and the right heatmap partial LOF events with high (20-50%) residual activity. The pathogenicity plot shows variants predicted to be damaging by SIFT, Mutassessor or PROVEAN in red. All data were extracted from the UMD TP53 mutation database (http://p53.fr/tp53-database). Transcriptional activity data were determined in a yeast-based reporter assay [10]. Cooperativity mutations at residues E180 and R181 are highlighted in orange. B Non-hotspot TP53 missense mutants retain substantial transcriptional activity. Shown is the range of transcriptional activity for somatic TP53 mutations grouped according to mutation frequency in cancer patients. Cooperativity mutations are highlighted separately in orange. C, D Range and distribution of transcriptional activity in p53 mutant cancer patients. In all box plots, boxes indicate median and interquartile range, whiskers the 10–90 percentile. Pie charts depict the percentage of variants (or tumors) falling into different categories of transcriptional activity as indicated. 31.6% of p53 variants and 27.6% of tumors with p53 missense mutations fall into the partial-LOF category defined by a residual transcriptional activity of 10–50%.