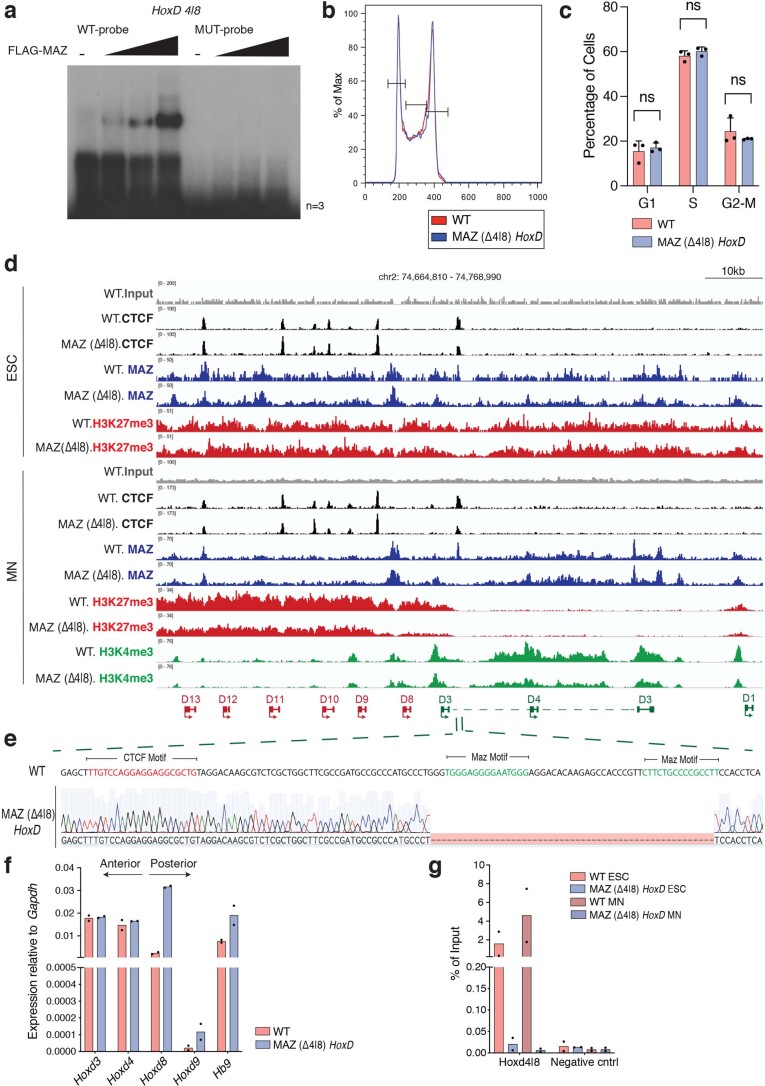
Extended Data Fig. 7. Loss of MAZ motif alters Hox gene expression and HoxD chromatin domains.
a, EMSA assay for FLAG-MAZ binding to MAZ motifs at Hoxd4|8 MAZ binding sites in WT probes in comparison to mutant (MUT) probes. One representative of three biological replicates is shown. b, FACS analysis of cell cycle in WT versus MAZ (∆4|8) ESCs (see Supplementary Fig. 7b for gating of cells). c, Quantification of cell cycle analysis by FACS in WT versus MAZ (∆4|8) ESCs (see Supplementary Fig. 7b for gating of cells). Results are represented as mean values and error bars indicating SD across three biological replicates. Two-sided Student’s t-test (unpaired) was used without multiple testing correction (black dots: individual data points). d, Normalized ChIP-seq densities for CTCF, MAZ, and indicated histone post-translational modifications (PTMs) in WT, and MAZ (∆4|8) ESCs and MNs in the HoxD cluster. ChIP-seq tracks are from one representative of two biological replicates for CTCF, and one replicate for MAZ and histone PTMs. e, MAZ binding site deletion via CRISPR is depicted for the 4|8 site at the HoxD cluster. f, RT-qPCR for the indicated Hox genes in the HoxD cluster in MNs upon MAZ (∆4|8). Results are represented as mean values of two independent experiments with individual data points overlaid. g, MAZ ChIP-qPCR analysis in the HoxD cluster in mESCs and MNs upon MAZ (∆4|8). Results are represented as mean values of two independent experiments with individual data points overlaid.