Fig. 7. Model of the role of MAZ and CTCF in genome structure and function.
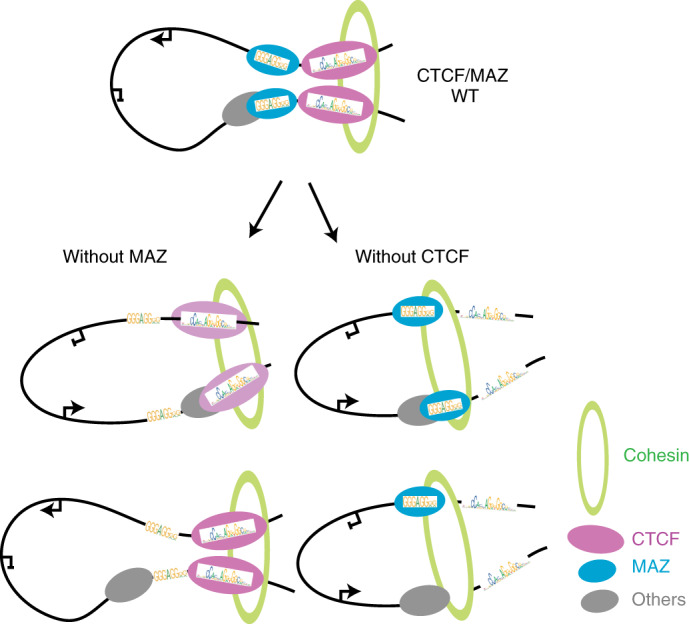
Proposed model for the effect of MAZ loss (left) and CTCF depletion (right) on loops, as well as the existing loop-extrusion model of CTCF and cohesin53,54. MAZ interacts with RAD21 and localizes to loop anchors similar to CTCF (top). In the absence of MAZ, some looping interactions are impacted, accompanied by decreased CTCF occupancy (bottom left). In the absence of CTCF, TADs and more looping interactions are disrupted44 (bottom right). Based on the regulatory interactions impacted and the underlying chromatin context, such topological changes can be accompanied by alterations in gene expression as evident at the identified Hox clusters. The relative orientations of MAZ and CTCF motifs are depicted based on analyses of their presence at loop anchors, as shown in Extended Data Fig. 10.
