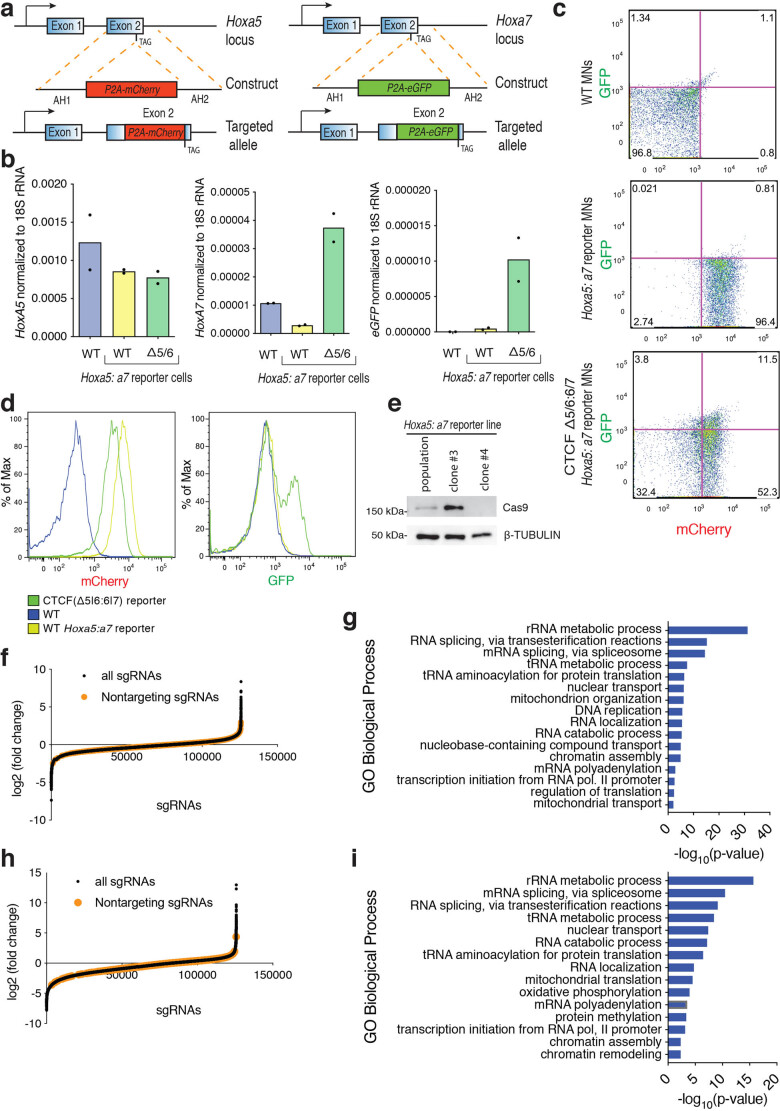
Extended Data Fig. 1. Genome-wide CRISPR KO screen shows loss of essential genes.
a, Strategy for generating the Hoxa5:a7 reporter mESC line via CRISPR. AH1, AH2: arm of homology 1, 2, respectively. b, RT-qPCR signal for Hoxa5, Hoxa7, and eGFP normalized to 18S ribosomal RNA in WT MNs, Hoxa5:a7 reporter MNs, and CTCF(Δ5|6) Hoxa5:a7 reporter MNs. MNs were obtained through in vitro differentiation of ESCs in two biological replicates. Δ5|6 denotes a CTCF binding site deletion between Hoxa5-6. c, FACS data showing mCherry and GFP reporter expression in WT MNs (top), Hoxa5:a7 reporter MNs (middle), and CTCF(Δ5|6:6|7) Hoxa5:a7 reporter MNs (bottom). These plots are one representative of three biological replicates. Percentage of positive population in each quadrant is indicated (see Supplementary Fig. 7a for gating of cells). d, Flow cytometry analysis of mCherry and GFP reporter expression in MNs with the indicated genotypes: WT, Hoxa5:a7 reporter, and CTCF(Δ5|6:6|7) reporter (see Extended Data Fig. 4 for RT-qPCR of Hox genes and Supplementary Fig. 7a for gating of cells). These plots are one representative of three biological replicates. Δ5|6:6|7 denotes 2 CTCF binding site deletions. e, Cas9 protein expression in dual-reporter mESCs transduced with Cas9 lentivirus and selected with blasticidin. β-TUBULIN serves as loading control. f, Fold change of sgRNAs in the primary screens comparing ESCs to plasmid library. g, Gene Ontology (GO) analysis of biological processes in negatively selected genes in ESCs compared to plasmid library (FDR < 0.05). PANTHER overrepresentation test tool was used for GO analysis. h, Fold change of sgRNAs in the primary screens comparing MN population to ESC population. i, GO analysis of biological processes in negatively selected genes in MNs compared to ESCs (FDR < 0.05). PANTHER overrepresentation test tool was used for GO analysis.