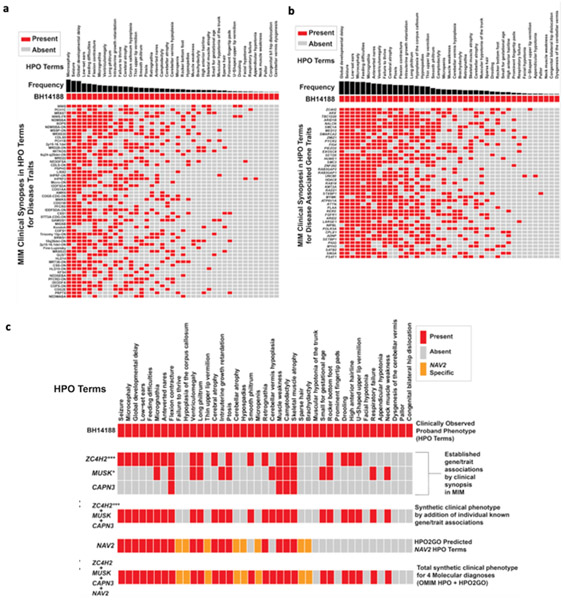
Figure 4. Annotation grids showing overlap of proband phenotypic traits to known disease traits and known disease gene-associated traits used to explore multilocus pathogenic variation (MPV).
a) Known Mendelian Inheritance of Man (MIM) disease match (p < 0.0037) annotation grid showing phenotypic overlap (red) with proband BH14188. Y-axis shows known diseases from OMIM/Orphanet. X-axis depicts phenotypes using HPO terminology. Frequency of disease-associated HPO terms is shown in black. b) Proband phenotypic overlap within the top (p < 0.01) matching HPO-term annotated gene-set. Matches are rank ordered by semantic similarity. Frequency of disease gene-associated HPO terms is shown in black. c) HPO analysis of terms from OMIM/Orphanet annotation to proband phenotype with known disease genes (ZC4H2, MUSK, and CAPN3) and novel candidate disease gene (NAV2) identified by research exome analysis. ZC4H2, MUSK, and CAPN3 were initially an analyzed individually, followed by the combined unique HPO term set of ZC4H2, MUSK, and CAPN3. NAV2 was initially analyzed individually using HPO2GO predicted terms, followed by the analysis of the totality of all 4 genes identified by research exome analysis (ZC4H2, MUSK, CAPN3, NAV2). Novel disease gene candidate NAV2 HPO2GO predicted phenotypic traits present in the proband are shown in orange. Asterisks represent significance according to standard convention (*** < 0.01, * < 0.05).