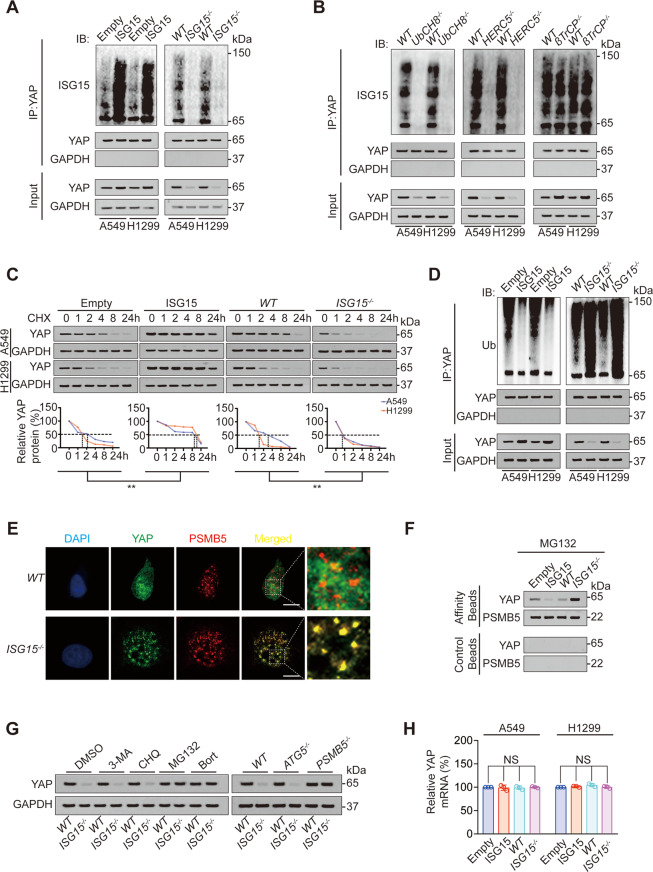
Fig. 1. YAP ISGylation inhibits its ubiquitination and increases its stability.
A Co-IP was performed in control, ISG15 overexpression or knockout A549 and H1299 cells using anti-YAP antibodies. The YAP level in each co-IP sample was adjusted to the same protein content. Indicated proteins were further analyzed by IB. B Co-IP was performed in control or UbCH8, HERC5, or βTrCP knockout A549 and H1299 cells using anti-YAP antibodies. The YAP level in each co-IP sample was adjusted to the same protein content. Indicated proteins were further analyzed by IB. C CHX (10 μg/ml) chase experiments were performed in control, ISG15 overexpression or knockout A549 and H1299 cells. The relative protein levels of YAP were shown as the ratios between YAP and GAPDH, and the “0 h” points were arbitrarily set to 100%. D Co-IP was performed in control, ISG15 overexpression or knockout A549 and H1299 cells using anti-YAP antibodies. The YAP level in each co-IP sample was adjusted to the same protein content. Indicated proteins were further analyzed by IB. E Co-localization of YAP and PSMB5 was analyzed in WT or ISG15−/− A549 cells before being photographed using a confocal microscope. Scale bar, 20 μm. F Association of YAP and PSMB5 analyzed by IB in proteasomes isolated from A549 cells with or without ISG15 overexpression or knockout in the presence of MG132 (8 μM, 24 h). Samples from affinity or control beads were analyzed in parallel. G YAP expression was analyzed by IB in WT or ISG15−/− A549 cells with 3-MA, CHQ, MG132, or Bort treatment or with ATG5 or PSMB5 knockout. H YAP mRNA level was analyzed in control, ISG15 overexpression or knockout A549, and H1299 cells. The data are shown as the mean ± SD from three biological replicates (including IB). Data in C were analyzed using a two-way ANOVA test. Data in H were analyzed using a one-way ANOVA test. NS nonsignificant.