Abstract
The SARS-CoV-2 (SARS-CoV-2) virus has caused a worldwide pandemic because of the virus's ability to transmit efficiently human-to-human. A key determinant of infection is the attachment of the viral spike protein to the host receptor angiotensin-converting enzyme 2 (ACE2). Because of the presumed zoonotic origin of SARS-CoV-2, there is no practical way to assess the susceptibility of every species to SARS-CoV-2 by direct challenge studies. In an effort to have a better predictive model of animal host susceptibility to SARS-CoV-2, we expressed the ACE2 and/or transmembrane serine protease 2 (TMPRSS2) genes from humans and other animal species in the avian fibroblast cell line, DF1, that is not permissive to infection. We demonstrated that expression of both human ACE2 and TMPRSS2 genes is necessary to support SARS-CoV-2 infection and replication in DF1 and a non-permissive sub-lineage of MDCK cells. Titers of SARS-CoV-2 in these cell lines were comparable to those observed in control Vero cells. To further test the model, we developed seven additional transgenic cell lines expressing the ACE2 and TMPRSS2 derived from Felis catus (cat), Equus caballus (horse), Sus domesticus (pig), Capra hircus (goat), Mesocricetus auratus (Golden hamster), Myotis lucifugus (Little Brown bat) and Hipposideros armiger (Great Roundleaf bat) in DF1 cells. Results demonstrate permissive replication of SARS-CoV-2 in cat, Golden hamster, and goat species, but not pig or horse, which correlated with the results of reported challenge studies. Cells expressing genes from either bat species tested demonstrated temporal replication of SARS-CoV-2 that peaked early and was not sustained. The development of this cell culture model allows for more efficient testing of the potential susceptibility of many different animal species for SARS-CoV-2 and emerging variant viruses.
Keywords: SARS-CoV-2, ACE2, TMPRSS2, Animal, Replication, Model
1. Introduction
The current COVID-19 pandemic is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) which was first reported in Wuhan, China in late 2019. This virus most probably has its ecological reservoir in bats, and transmission of the virus to humans has likely occurred through an intermediate animal host which has not yet been identified (Conceicao et al., 2020; Damas et al., 2020). Coronaviruses (CoVs) are a large family of viruses, several of which cause respiratory diseases in humans, from mild disease with common cold-like symptoms to more serious diseases such as the Severe Acute Respiratory Syndrome (SARS) and the Middle East Respiratory Syndrome (MERS), both of which have high case fatality rates and were detected for the first time in 2002 and 2012, respectively.
CoVs are enveloped, single-stranded, positive-sense RNA viruses that belong to the subfamily Orthocoronavirinae within the family Coronaviridae, Order Nidovirales. The viruses are divided into four genera: alpha-, beta-, gamma- and delta-CoV based on phylogenetic and genomic structure (Ksiazek et al., 2003; Li et al., 2019). All CoVs currently known to cause disease in humans belong to the alpha- or beta-CoV groups (Brian and Baric, 2005; Cui et al., 2019). In addition, alpha-CoV, beta-CoV and gamma-CoV induce significant disease in various domestic animal species, including porcine transmissible gastroenteritis virus, porcine enteric diarrhea virus (PEDV), swine acute diarrhea syndrome coronavirus (SADS-CoV), and infectious bronchitis virus (IBV) in poultry (Brian and Baric, 2005; Cavanagh, 2007; Cui et al., 2019; Lin et al., 2016; Zhou et al., 2018). Based on sequence analysis, human coronaviruses have animal origins. The SARS-CoV, MERS-CoV, HCoV-NL63 and HCoV-229 E are thought to have originated in bats, whereas HCoV-OC43 and HKU1 appear to have come from rodents (Forni et al., 2017). With the 2002 SARS-CoV-1, the virus was also detected in civet cats and was suggested as an intermediary host to humans, and the 2012 MERS-CoV appeared to have spread from bats to dromedary camels and then to humans (Han et al., 2015; Muller et al., 2014; Song et al., 2005).
The main surface protein of CoVs is the spike (S) protein that facilitates receptor binding and fusion of the viral lipid envelope with the host cell membrane. Receptor binding is facilitated by the S1 subunit while the S2 subunit is involved with fusion of the viral membrane with the cell membrane (Hoffmann et al., 2020b; Hulswit et al., 2016). For these two events to occur, the S protein needs to be post-transitionally modified by two different host proteases to become activated. First, furin-like proteases cleave the S protein at the S1/S2 site that contains a multiple basic amino acid motif that is different from SARS-CoV (Hoffmann et al., 2020a). The S protein undergoes additional cleavage at the S2’ site by the cellular type II transmembrane serine protease, TMPRSS2 (Glowacka et al., 2011; Matsuyama et al., 2010; Shulla et al., 2011). However, other proteases have been described to activate CoVs including cathepsin L, TMPRSS11A, TMPRSS11D, TMPRSS4, TMPRSS13 and furin (Bestle et al., 2020; Heald-Sargent and Gallagher, 2012; Hoffmann et al., 2020a, 2021; Millet and Whittaker, 2015; Zang et al., 2020; Zmora et al., 2018).
SARS-CoV and SARS-CoV-2 utilize the angiotensin-converting enzyme 2 (ACE2) as the receptor for attachment on host cells with the S protein (Hoffmann et al., 2020b). ACE-2 is a single-pass type I transmembrane protein, with its enzymatically active domain exposed on the surface of cells in lungs and other tissues. ACE2 catalyzes the conversion of angiotensin I into angiotensin 1-9 and angiotensin II into angiotensin 1-7, which are involved with vasodilation effects in the cardiovascular system (Feng et al., 2008; Patel et al., 2016). Due to conservation of the ACE2 gene among animal species, the potential host range of SARS-CoV-2 is thought to be extensive.
The ACE2 and TMPRSS2 genes have homologues in many animal species (Bestle et al., 2020; Damas et al., 2020). Several species, including house cats, ferrets, and golden hamsters, have been shown to be naturally and/or experimentally infected with SARS-CoV-2 (Ghai et al., 2021). These three species have >80% sequence similarity in their ACE2 and TMPRSS2 genes when compared to the human genes. The chicken, which does not appear to be a susceptible host, has an ACE2 homology of less than 70% to the human gene (Suarez et al., 2020). However other species like pigs have a sequence similarity of >80%, but are poorly susceptible to infection (Meekins et al., 2020; Pickering et al., 2021; Schlottau et al., 2020). Based on previous work with SARS-CoV, the binding of S1 to ACE2 can be defined by the interaction of relatively few amino acids, and predictions of host susceptibility based on these interactions have been made (Damas et al., 2020; Liu et al., 2020). Despite the clear importance of the binding of the spike protein to ACE2, the prediction of host susceptibility does involve other factors including the level and tissue distribution of ACE2 expression and the requirement for protease activation.
Because chickens are not susceptible to SARS-CoV-2 virus, and their ACE2 and TMPRSS2 protease are distinctly different from the human equivalents, we developed an avian cell line to screen the potential host range of infection of the virus through the expression the ACE2 and TMPRSS2 genes from human and animal species to provide novel insights into the receptor usage, replication and potential host range of SARS-CoV-2. These studies were designed to determine if the host restriction is strictly from the difference in the receptor and/or protease. One long-term goal of this work is to develop a predictive framework for improved epidemic surveillance to include protection of agriculturally relevant species and animal species that are hard to test experimentally.
2. Materials and methods
Viruses. The USA-WA1/2020 (BEI NR-58221 isolate of SARS-CoV-2 (SARS-CoV-2) was obtained from BEI Research Resources Repository, National Institute of Allergy and Infectious Diseases, National Institutes of Health (Gralinski and Menachery, 2020). Experiments with SARS-CoV-2 were performed in a biosafety level-3 enhanced facility with procedures approved by the U.S. National Poultry Research Center Institutional Biosafety Committee.
Cell lines. DF1 (avian fibroblast), Madin-Darby Canine Kidney (MDCK) and Vero (African Green monkey kidney, CCL-81) cells were seeded and propagated with standard procedures for adherent cells in flasks containing Dulbecco's Modified Eagle Medium (DMEM) (ThermoFisher Scientific, Waltham, MA) with 10% Fetal Bovine Serum (Sigma Chemical Company, St. Louis, MO) and 1% Antimicrobial-Antimycotic (GeminiBio, Sacramento, CA). MDCK cells were obtained from ATCC and were included because this sub-lineage was not able to support SARS-CoV-2 replication (Hoffmann et al., 2020b). Vero cells were obtained from the International Reagent Resource (FR-243).
Construction of transgenic cell lines using lentivirus vectors expressing human ACE2 and TMPRSS2. DF1 and MDCK cells were seeded at a density of 0.5 × 105 in 500 μl DMEM containing 10% Fetal Bovine Serum and 1% Antimicrobial-Antimycotic (Sigma), in one well each of a 12 well plate, and left overnight as above. The lentivirus contained the human ACE2 gene under control of the CMV promoter along with green fluorescent protein (GFP) also under control of a separate CMV promoter with transduction carried out according to the manufacturers recommendation (Origene Technologies, Rockville, MD). For TMPRSS2 transduction, lentivirus particles containing the human TMPRSS2 gene under control of the CMV promoter and red fluorescent protein (RFP) gene under control of a separate CMV promoter (Gentarget, San Diego, CA). Transduction was confirmed using an EVOS 5000 (Invitrogen, Carlsbad, CA), equipped with GFP, RFP, DAPI and transmitted light cubes, to visualize cells expressing GFP or RFP, or both. Production of DF1 or MDCK cells expressing only human ACE2 (defined as + -) or only human TMPRSS2 (defined -+), or both (defined as ++), was confirmed by RT-PCR and purification by FACS cell sorting for either green or red fluorescence. For construction of cells expressing both, the human ACE2 was first inserted and purified for GFP (99% GFP-positive) followed by human TMPRSS2 insertion and cell sorting for both RFP- and GFP-positive cells (See Supplemental Fig. 1). Confirmation of human ACE2 and human TMPRSS2 expression was performed by RT-PCR and Western blot.
Construction of transgenic DF1 cell lines expressing different animal ACE2 and TMPRSS2 genes using the PiggyBac transposon vector. GenBank accession numbers used to construct all species plasmids can be found in Supplemental Table 1. The ACE2 and TMPRSS2 genes from cat (Felis catus), horse (Equus caballus), domestic pig (Sus domesticus), goat (Capra hircus), Golden hamster (Mesocricetus auratus), Little Brown bat (Myotis lucifugus) and Great Roundleaf bat (Hipposideros armiger) were de novo synthesized into the PiggyBac® transposon expression plasmids under control of the CMV promoter (VectorBuilder Inc., Chicago, IL). As with the human genes, GFP was included for ACE2 detection and purification, and RFP was included for TMPRSS2 detection and purification. Frozen E. coli plasmid glycerol stocks, containing either ACE2 or TMPRSS2, were streaked onto LB agar plates (Invitrogen) containing 100 μg/mL of Carbenicillin (Sigma). Single colonies were selected and incubated in 50 mL LB Broth, containing 100 μg/mL of Carbenicillin, with gentle agitation overnight in an incubator/shaker at 34 °C (Amerex Instruments). Plasmid DNA was isolated per the manufacturers recommendation.
PiggyBac Transfection with animal ACE2 and TMPRSS2. DF1 cells were transfected with plasmids containing animal ACE2 using Lipofectamine 3000 (Invitrogen) according to the manufacturer's protocol. Transposase and Transposon DNA were added at 1:1 ratio in 10% FBS. Cells were incubated for 72 h at 39 °C, after which expression was confirmed using an EVOS 5000 as above. Cells were purified for GFP expression (>99%) using flow cytometry as below and those cells seeded as above for transfection with plasmid constructs containing the TMPRRS2 and RFP genes. Cells were purified by FACS cytometry for GFP and RFP (>99%) and used for infection studies.
Fluorescent-activation cell sorting (FACS). Transgenic cells expressing ACE2, TMPRSS2 or both, were grown to 90% confluence, trypsinized, pelleted by centrifugation (1500×g for 10 min at room temperature) and strained through a 50 μm cell strainer (Fisher Scientific). Cells were sorted for GFP or RFP, or both, at the University of Georgia (Athens, Georgia), Flow Cytometry Core Center, using a Beckman Coulter Moflo Astrios EQ (Beckman Coulter).
RNA extraction and RT-PCR for human ACE2 and TMPRSS2. Total RNA was extracted from 2.5 × 105 cells in 1 well of a 6 well plate from Vero, DF1, DF1 +-, DF1 -+, DF1 ++, MDCK, MDCK + -, MDCK -+ and MDCK ++ as previously described (Jiang et al., 2011). Final RNA extraction was carried out using the ZYMO Direct-zol Mini-Prep Plus Kit (Zymo Research, Irvine, CA) per manufactures instructions.
Superscript 4 Reverse Transcriptase (Invitrogen) was used according to manufacturer's instructions. One μl of 2 μm gene specific primer and 11 μl of RNA were used for all reactions. Gene specific first strand primers as designated in Supplementary Table 2. Two μl of cDNA template was used for all cell lines. Reactions were conducted using NEB Phusion Hi Fi Polymerase (New England Biolabs, Ipswich, MA). Reactions were comprised of 4 μl 5X Phusion Buffer, 0.4 μl 10 mM DNTPs, 1 μl of Forward and Reverse Primer, 2 μl of cDNA, 0.6 μl of DMSO, 0.2 μl of DNA polymerase, and 11 μl of ultrapure water (Invitrogen). Primers used for human ACE2 PCR are found in Supplementary Table 2. Reaction conditions were 98 °C for 30 s, followed by 35 cycles of 98° for 10 s, 68 °C for 30 s and 72 °C for 1 min, after which a final extension of 10 min at 72° was added.
Primers for human TMPRSS2 are found in Supplementary Table 2. Annealing temperature for reactions was 66 °C and all other conditions were identical to human ACE2. PCR products were visualized on 1% agarose gel (Bio-Rad Laboratories, Hercules, CA) containing SYBR Safe (Invitrogen) using a documentation system (Syngene International Ltd, Bengaluru, India).
RNA extraction and RT-PCR for animal species ACE2 and TMPRSS2. Total RNA was extracted as above (Jiang et al., 2011). Superscript 4 Reverse Transcriptase (Invitrogen) was used according to manufacturer's instructions. One μl of 2 μm gene specific primer and 11 μl of RNA were used for all reactions. The gene specific first strand and RT-PCR primers used are found in Supplementary Table 2 Two μl of cDNA template was used for all cell lines. Reactions were conducted using NEB Phusion Hi Fi Polymerase (New England Biolabs, Ipswich, MA). Reactions were comprised of 4 μl 5X Phusion Buffer, 0.4 μl 10 mM DNTPs, 1 μl of Forward and Reverse Primer, 2 μl of cDNA, 0.6 μl of DMSO, 0.2 μl of DNA polymerase, and 11 μl of ultrapure water (Invitrogen). Primers used for animal species ACE2 and TMPRSS2 are found in Supplemental Fig. 2. Reaction conditions for ACE2 were 98 °C for 30 s, followed by 35 cycles of 98° for 10 s, 68 °C for 30 s and 72 °C for 1 min, after which a final extension of 10 min at 72° was added. Annealing temperature for TMPRSS2 reactions were 66 °C and all other conditions were identical to animal ACE2. PCR products were visualized on 1% agarose gel (Bio-Rad Laboratories, Hercules, CA) containing SYBR Safe (Invitrogen) using a documentation system (Syngene International Ltd, Bengaluru, India).
Detection of human ACE2 and TMPRSS2 protein expression by Western blot, and immunohistochemistry to detect SARS-CoV-2. Western blot detection was performed as described (Kapczynski et al., 2016). The blot was then incubated overnight at 4 °C in primary antibody diluted 1:1500 in TBS. Primary monoclonal antibodies included mouse anti-human ACE2 (Origen), rabbit anti-human TMPRSS2 (Abcam, Cambridge, UK) and mouse anti-beta actin (Invitrogen). The blot was washed as before, incubated for 1 h, at room temperature, in secondary antibody diluted 1:20,000 in TBS with gentle rocking. Secondary antibodies included rat anti-mouse IgG1 HRP (Southern Biotech, Birmingham, AL), and mouse anti-rabbit IgG1 HRP (Southern Biotech). After incubation, the blot was washed 3 times as above in TBST. Pierce ECL substrate (Fisher) was added to the blot for 1 min and excess was removed by gentle wicking. The blot was placed into an x-ray cassette and exposed to x-ray film (Fisher) for 1 min, developed and fixed (Kodak).
For immunohistochemistry of SARS-CoV-2 replication, cells were seeded into an I-Bidi 8-well chambered slide (Fisher) at a density of 4 × 104 in 500 μl DMEM containing 10% FBS and grown overnight as above. When cells reached 75% confluence the media was removed, and virus was added at MOI of 1 as above. After 48 h, the media was removed and cells were fixed for 5 min at 4C in 1:1 ice cold ethanol:methanol. Cells were then washed twice with cold PBS as above. Cells were blocked as above for 1 h at room temperature then washed 3 time with TBS. Primary antibodies against SARS-CoV-2 included rabbit anti-Nucleoprotein MAb (Origene) and rabbit anti-Spike MAb (Origene), diluted as above, were added for 1 h at room temperature. Cells were washed 3 times with PBS and incubated in the secondary antibody, goat anti-rabbit IgG H&L (Alexa Fluor® 555) (ABCAM) diluted 1:20,000 in TBS, for 1 h at room temperature. Cells were then washed 3 times with PBS and counterstained with DAPI (Invitrogen) for 5 min. Cells were washed 3 times with PBS then allowed to air dry. Once dry, cells were mounted with ProLong™ Gold Antifade Mountant (Fisher) and sealed with glass coverslips after 24 h. Immunofluorescence was visualized with an EVOS 5000 (Invitrogen).
Comparison of SARS-CoV-2 replication dynamics among cell lines. Cell lines were tested for virus replication by inoculating them with SARS-CoV-2 at an MOI of 1 added directly when cells were approximately 70–90% confluent in 6 well plates. For each cell line, media was removed from three wells and 0.4 mL of virus was added. The same volume of sterile medium was added to wells on each plate to serve as a sham inoculated control. The plates were incubated for 1 h at 37 °C, 5% CO2 to allow virus to adsorb to the cells. Each well was washed 3-times with sterile PBS prewarmed at 37 °C to remove unbound virus. Finally, 3 mL growth medium was added to each well and the cells were incubated at 37 °C with 5% CO2. Supernatant (0.2 mL) was collected from each well individually at 6, 12, 24, 36, 48 and 72 h post inoculation (hpi) for detection of replicating virus by RT-PCR, and detection of cytopathic effect. After 72 hpi, plates were frozen and thawed at −80C (3x total) and 400 μl of cell culture supernatant was transferred onto fresh cell cultures as above for a pass 2 to confirm infectious virions were produced in the avian cells.
Quantitative real-time RT-PCR to detect SARS-CoV-2. Quantitative RT-PCR was utilized to detect and determine virus titers in cell culture supernatants. RNA was extracted with the Ambion Magmax kit (ThermoFisher). The US Centers for Disease Control N1 primers and probe for SARS-CoV-2 were used with the AgPath ID one-step RT-PCR kit (Lu et al., 2020). The cycling conditions for the RT step were modified to accommodate the recommended kit conditions. A standard curve of RNA from titrated SARS-CoV-2 virus stock was run in duplicate to establish titer equivalents of virus and the viral titer was extrapolated from the standard curve.
TMPRSS2 genetic analysis. TMPRSS2 gene sequences from animal species were obtained from GenBank. Sequences were aligned with Clustal V (Lasergene 10.0, DNAStar, Madison, WI), and protein architecture derived from The National Center for Biotechnology (www.ncbi.nih.gov).
Statistical analysis. Viral titers at 48 hpi were compared with the two-way ANOVA with Tukey multiple comparison (Prism 9.1.0 GraphPad Software, San Diego, CA). Different lower case letters indicate statistical significance between compared groups. All statistical tests used P < 0.05 as being statistically significant.
3. Results
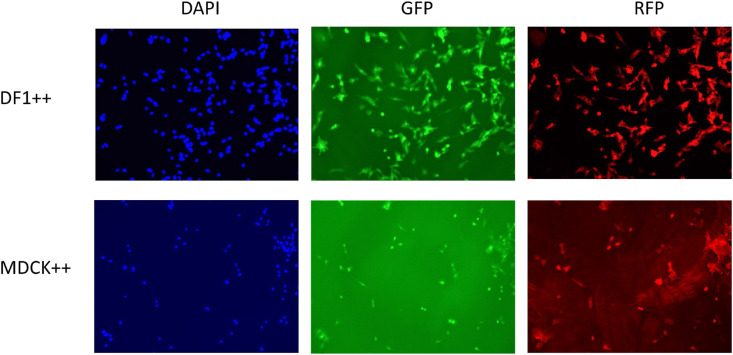
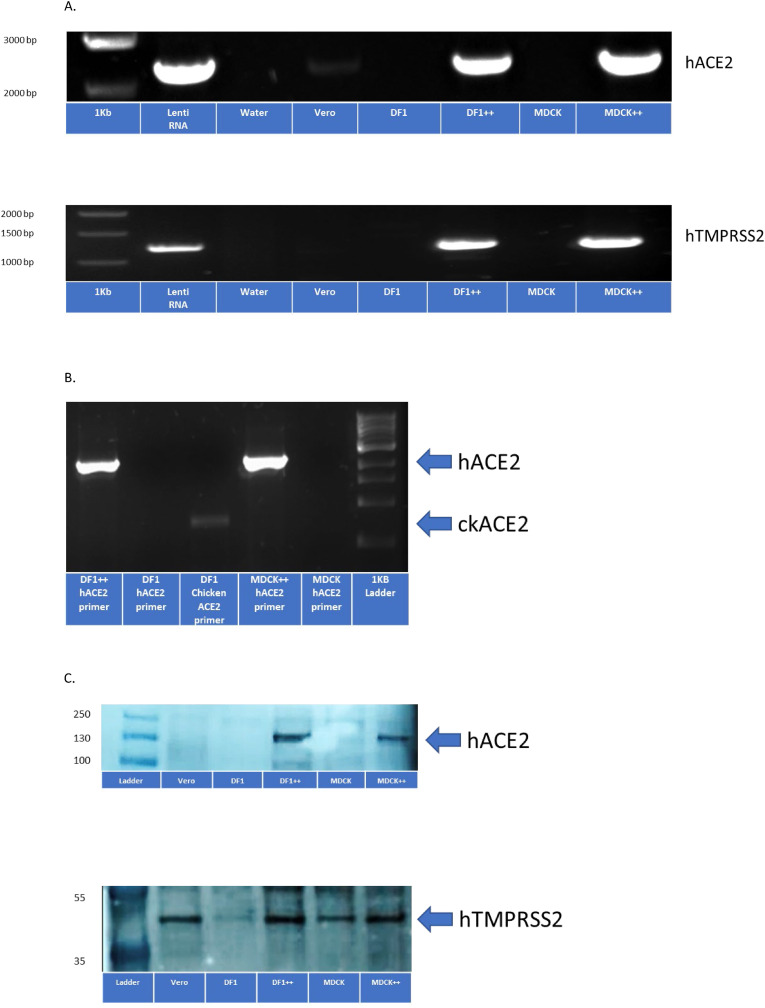
Development of DF1 and MDCK cell lines expressing human ACE2 and TMPRSS2. These studies were designed to transgenically introduce the human receptor and protease used by SARS-CoV-2 into the avian non-permissive cell line, DF1, and a MDCK cell line to test requirements for replication competence and establish a model for infection potential. A lentivirus approach was used to deliver the human ACE2 and human TMPRSS2 genes, under control of the CMV promoter. The lentivirus constructs co-expressed GFP (ACE2) and/or RFP (TMPRSS2) to allow FACS sorting for purification of cells containing each target gene or both genes (Supplemental Figs. 1A, B, C). Positive DF1 and MDCK cells were demonstrated expressing either the human ACE2 gene or human TMPRSS2 gene alone, or both, based on microscopy and two-color cell sorting (Fig. 1 , Supplemental Fig. 2). Detection of the inserted genes was confirmed with RT-PCR using primers specific for the human and chicken genes (Fig. 2 A and B). Expression of human ACE2 and human TMPRSS2 protein in DF1 ++ and MDCK ++ cells was confirmed via Western blot (Fig. 2C). There was detection of the canine TMPRSS2 by the human TMPRSS2 MAb in MDCK untransformed cells, but the avian TMPRSS2 was not detected in untransformed DF1 cells with this antibody.
Fig. 1.
DF1 and MDCK cells expressing the human ACE2 (with GFP marker) and TMPRSS2 (with RFP marker) genes. DF1 and MDCK cells were transduced with lentivirus containing the human ACE2 gene and cells were FACS purified based on GFP expression. Lentivirus containing the humanTMPRSS2 gene was then transduced into the human-ACE2 expressing DF1 and MDCK cells. Following two-color FACS for GFP and RFP expressing cells, dual positive cells were grown for 48 h in an 8-chamber glass slide. Fluorescence was captured on an EVOS M5000 with added DAPI nuclear stain (blue) GFP and RFP.
Fig. 2.
Detection of human ACE2 and human TMPRSS2 expression in DF1 + + and MDCK + + cells. (A) DF1, DF1 ++, MDCK, MDCK ++ and Vero cells were grown at 37C in 5% CO2. After 72 h, RNA was extracted and primers specific for human ACE2 and human TMPRSS2 were used with RT-PCR to confirm expression in DF1 ++ and MDCK ++ cell lines. (B) Differential expression of human and chicken ACE2 in DF1, DF1 ++, MDCK, and MDCK ++ cell lines with primers specific for both. (C) Fifteen micrograms of protein were extracted from each cell line and separated by SDS-PAGE. Following transfer to nitrocellulose, membranes were probed by Western blot using rabbit monoclonal antibodies to the human ACE2 and TMPRSS2 proteins.
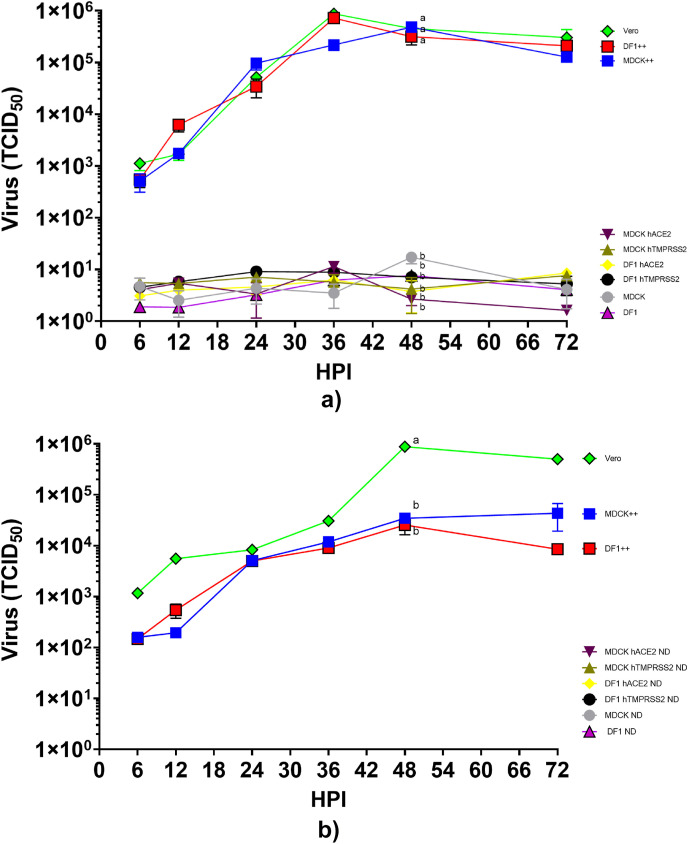
Comparison of SARS-CoV-2 replication dynamics in DF1 and MDCK cell lines expressing human ACE2 and/or TMPRSS2. Growth curves for all cell lines, Vero, DF1, and MDCK, and DF1 or MDCK expressing only human ACE2 (+-), only human TMPRSS2 (-+), or both (++) are shown in Fig. 3. No increase in virus titer was demonstrated in wild type DF1 or MDCK, or the DF1 and MDCK cells expressing single gene constructs with human ACE2 or human TMPRSS2 (Fig. 3A). In contrast, virus replication was observed in Vero (positive control), and both cell lines expressing both genes, DF1++ and MDCK ++. Virus growth was exponential until approximately 36–48 h post infection and was statistically higher in these cells than others tested at this time. Virus titers reached similar levels of approximately 105.6 TCID50 in these three cell lines, and demonstrated a requirement for expression of both the receptor and the protease. We next passaged the 72 h sample from each cell line after a freeze thaw cycle onto a subsequent plate of the same cells to determine if infectious virus could be passaged (Fig. 3B). No evidence of increased replication was seen on the second passage in any of the cell lines. The Vero, DF1++, and MDCK ++ passage 1 samples contained enough virus to induce infection and replication on passage 2, although the growth curves displayed a more linear increase in virus titer over time compared to passage 1 inoculated cells.
Fig. 3.
Growth of SARS-CoV-2 on DF1 and MDCK cells expressing either human ACE2, human TMPRSS2, or both (++). (A) DF1, DF1 expressing human ACE2, DF1 expressing human TMPRSS2, DF1 expressing both human ACE2 and TMPRSS2 (++), MDCK, MDCK expressing human ACE2, MDCK expressing human TMPRSS2, MDCK expressing both human ACE2 and TMPRSS2 (++), and Vero cells were inoculated with SARS-CoV-2 at multiplicity of infection (MOI) of 1. At time points indicated, supernatant samples were taken for RNA extraction and determination of viral titers by RT-PCR. The values shown are mean±standard deviation of triplicate samples. Two-way analysis of variance with Tukeys multiple comparison test was performed on titers at 48 h post inoculation to determine the statistical difference in virus titer between the cell lines. Lines with different lowercase letters indicate differences (p < 0.05). (B) Pass 2 of virus from cell culture lines expressing human ACE2, TMPRSS2, or both. After 72 h of growth, supernatants of pass 1 were transferred onto fresh monolayers of cells, allowed to absorb for 1 h and removed. Fresh media was added and samples were taken at time points indicated to determine virus titer by RT-PCR. Statistical analysis was performed at 48 h post inoculation. ND=Not detected.
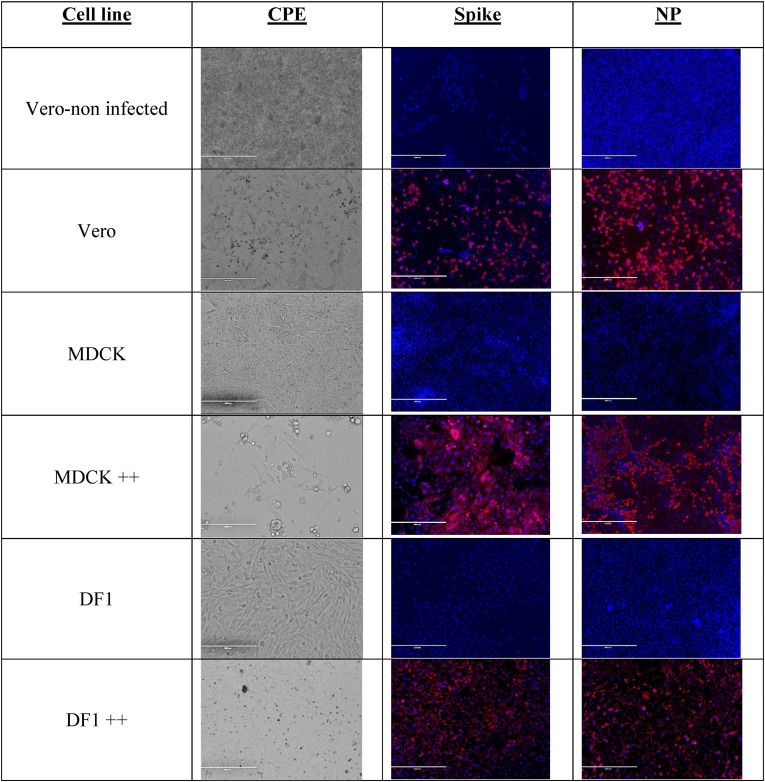
Comparison of cytopathic effects and detection of virus in cell lines expressing human ACE2 and TMPRSS2. The appearance of CPE and confirmation of virus protein inside of the cell lines was performed via light microscopy and immunohistochemistry with antibodies against the SARS-CoV-2 spike and nucleoprotein. Neither CPE nor virus could be detected in cells without virus (Fig. 4 ) or in the DF1 and MDCK inoculated cells. Likewise, cell lines containing the singular insertion of either the human ACE2 or TMPRSS2 did not exhibit CPE or positive viral staining (data not shown). Vero, DF1++, and MDCK++ demonstrated syncytia formation with loss of cell confluence. The monolayer also deteriorated by 72 hpi and CPE correlated with detection of high levels of expression of the viral spike and nucleoprotein by immunostaining at 48 hpi.
Fig. 4.
SARS-CoV-2-induced cytopathic effect and viral detection by immunohistochemistry in cells expressing human ACE2 and TMPRSS2. Vero, DF1, DF1 expressing both human ACE2 and TMPRSS2 (++), MDCK, and MDCK expressing both human ACE2 and TMPRSS2 (++) were grown at 37C in 5% CO2 on glass chamber slides. Cells were inoculated with SARS-CoV-2 at MOI of 1. At 48 h post inoculation monolayers were examined for cytopathic effect and detection of virus with rabbit monoclonal antibodies against SARS-CoV-2 spike and nucleoprotein. Cells were washed 3 times with PBS and incubated in the secondary antibody, goat anti-rabbit IgG H&L (Alexa Fluor® 555) for 1 h at room temperature. Cells were then washed counterstained with DAPI. Immunofluorescence was visualized with an EVOS 5000.
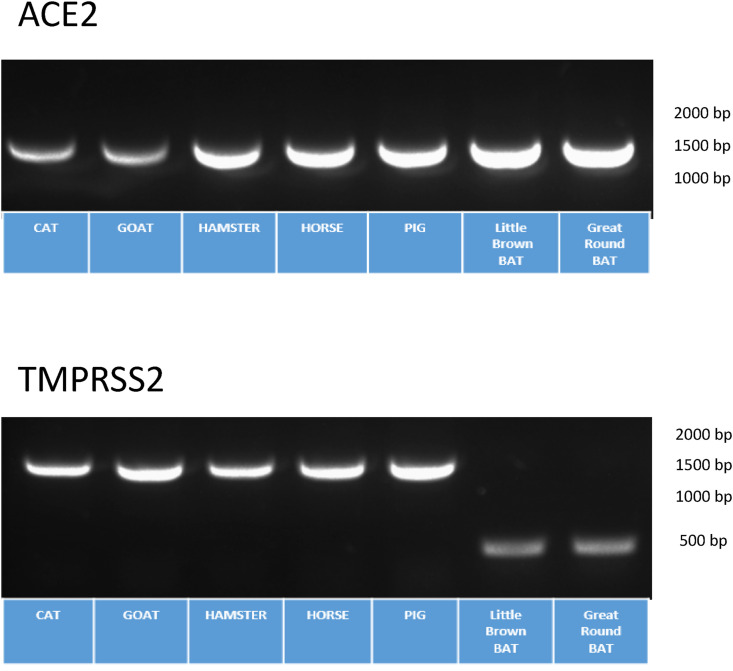
Development of cell lines expressing ACE2 and TMPRSS2 from different animal species. Having demonstrated a model of virus replication in the non-permissive avian DF1 cell line with insertion of the human ACE2 and TMPRSS2 genes, we next developed cells lines expressing other species ACE2 and TMPRSS2 to screen for potential animal hosts that could support replication. The ACE2 and TMPRSS2 genes from cat, goat, golden hamster, horse, pig, Little Brown bat, and Great Roundleaf bat were de novo constructed in the PiggyBac transposon system and transfected into DF1 cells. Purification of cells with green/red fluorescence was used as with the lentivirus system. As demonstrated in Fig. 5 , RT-PCR confirmed expression of animal ACE2 and TMPRSS2 in DF1 cells from FACS-sorted cells.
Fig. 5.
Transgenic DF1 cells expressing different animal species ACE2 and TMPRSS2 genes. (A) DF1 cells were transfected with PiggyBac® plasmid containing the ACE2 and TMPRSS2 genes from house cat (Felis catus), horse (Equus ferus), domestic pig (Sus domesticus), goat (Capra aegagrus), Golden hamster (Mesocricetus auratus), Little Brown bat (Myotis lucifugus) and Great Roundleaf bat (Hipposideros armiger). Cells were first created with the animal ACE2 gene and FACS purified based on GFP expression. The animal TMPRSS2 gene was then transfected into the DF1 cells expressing the animal ACE2 gene. Two-color FACS was performed based on GFP and RFP expression. Transgenic cells expressing animal ACE2 and TMPRSS2 were grown at 37C in 5% CO2. After 72 h, RNA was extracted and primers specific for the animals ACE2 and animal TMPRSS2 were used with RT-PCR to confirm animal species ACE2 and TMPRSS2 expression in DF1 cells.
3.1. SARS-CoV-2 replication in cells expressing animal ACE2 or TMPRSS2
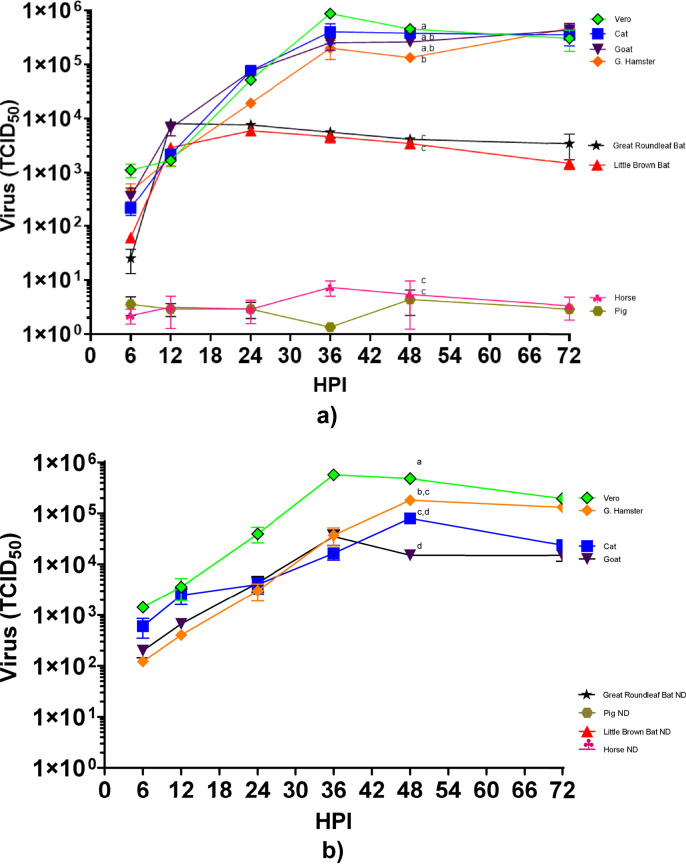
The replication kinetics of SARS-CoV-2 virus in DF1 cell lines expressing the ACE2 and TMPRSS2 genes from the different animal species was determined. Results demonstrate that the SARS-CoV-2 virus could replicate to high levels in DF1 cell lines expressing the ACE2 and TMPRSS2 genes from cat, goat and golden hamster (Fig. 6 A). Virus titers reached similar levels of approximately 105.1 to 105.8 TCID50 at 36 h post infection in these lines, which was similar to that observed in the Vero control cells. No virus replication was observed in the cells expressing the receptor and protease from pig or horse species. Both bat species demonstrated initial gains in virus titers, between 103.3 and 103.9 TCID50 at 12 h post infection that did not increase after this time. The 72 hpi sample from all cell lines were passaged onto a subsequent plate of cells. Passage 2 results indicate viral infection and replication from plates containing the cat, goat and golden hamster animal cell lines (Fig. 6B). As observed previously, a linear shaped curve in virus replication was observed in passage 2. Neither the pig nor the horse cell lines had evidence of virus replication in passage 2. The samples from the two bat species cell lines also had no evidence of replication on passage 2.
Fig. 6.
Growth of SARS-CoV-2 in DF1 cells expressing ACE2 and TMPRSS2 from different animal species. (A) DF1 cells expressing cat, horse, pig, goat, Golden hamster, Little Brown bat, and Great Roundleaf bat were inoculated with SARS-CoV-2 at multiplicity of infection (MOI) of 1. At time points indicated, supernatant samples were taken for RNA extraction and determination of viral titers with RT-PCR. The values shown are mean±standard deviation of triplicate samples. Two-way analysis of variance with Tukeys multiple comparison test was performed on titers at 48 h post inoculation to determine the statistical difference in virus titer between the cell lines. Lines with different lowercase letters indicate differences (p < 0.05). (B) Pass 2 of virus from cell culture lines animal species ACE2 and TMPRSS2. After 72 h of growth, supernatants of pass 1 were transferred onto fresh monolayers of cells, allowed to absorb for 1 h and removed. Fresh media was added and samples were taken at time points indicated to determine virus titer with RT-PCR. Statistical analysis was performed at 48 h post inoculation. ND=Not detected.
Sequence analysis of available TMPRSS2 sequence data for human and animal species demonstrated a truncation at the 5’ end of the bat protein compared to human or other animals (Supplemental Fig. 3). The human protein has 492 amino acids (AA), whereas the Little Brown bat contains 243 AA and Great Roundleaf bat has 384 AA. It is not clear and remains to be determined if the bat sequences available in GenBank were incorrectly annotated and are not representative of the complete protein, and that the bat species TMPRSS2 tested here may not be functional due to the missing the N-terminal portion of the protein. The Little Brown bat open reading frame begins at human amino acid position 255, and the Great Roundleaf bat begins at human position 113. Interestingly, Brandts bat (Myotis brandtii) contained a protease similar to human and other animals.
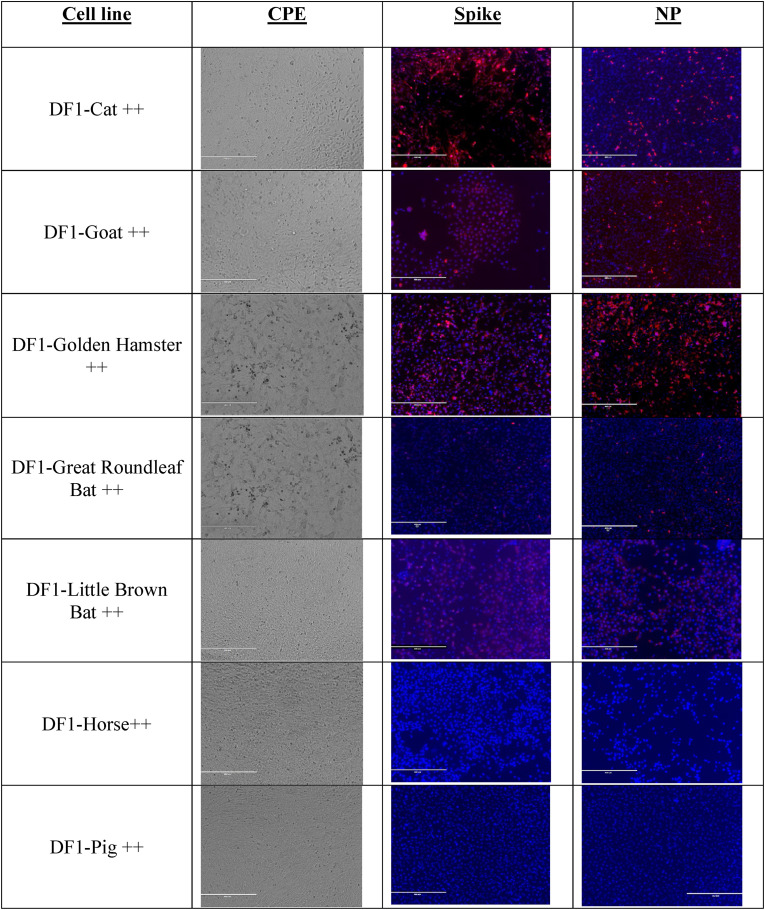
Comparison of cytopathic effects and detection of virus in cell lines expressing animal ACE2 and TMPRSS2. As before, detection of virus was observed via CPE and immunostaining of transgenic cell lines. As demonstrated in Fig. 7 , we detected cytopathic effects in cell lines that supported growth of the virus, including the ones expressing the cat, goat and golden hamster genes. We also observed CPE in both the cell lines expressing the bat genes that appeared more rapidly in the Great Roundleaf bat cell line compared to the Little Brown bat cell line. Staining for viral proteins was greatest in cells expressing cat, goat or golden hamster transgenes. Interestingly, we did observe positive staining in the bat species cells; however, it was visibly reduced compared to the other positive cell lines. We did not observe either CPE or viral staining in the cell lines expressing pig and horse genes. As expected, CPE and viral staining were observed in passage 2 in cat, goat, golden hamster cell lines but not bat species (data not shown).
Fig. 7.
SARS-CoV-2 induced cytopathic effect and viral detection by immunohistochemistry in DF1 cells expressing animal species ACE2 and TMPRSS2. DF1 cells expressing animal ACE2 and TMPRSS2 were grown at 37C in 5% CO2 on glass chamber slides. Cells were inoculated with SARS-CoV-2 at MOI of 1. At 48 h post inoculation monolayers were examined for cytopathic effect and detection of virus with rabbit monoclonal antibodies against SARS-CoV-2 spike and nucleoprotein. Cells were washed 3 times with PBS and incubated in the secondary antibody, goat anti-rabbit IgG H&L (Alexa Fluor® 555) for 1 h at room temperature. Cells were then washed counterstained with DAPI. Immunofluorescence was visualized with an EVOS 5000.
4. Discussion
Several cell lines and organoids are currently in use or have been developed to study SARS-CoV-2 replication. Besides Vero cells, Caco-2, Calu-3, and Hek293 T cells, human lung, kidney, liver and blood vessel organoids have been demonstrated to be permissive for virus growth (Han et al., 2021; Harcourt et al., 2020; Kim et al., 2020; Lamers et al., 2020; Monteil et al., 2020; Ou et al., 2020; Suzuki et al., 2021; Zhao et al., 2020; Zhou et al., 2020). However, because these systems can naturally be infected, they are not useful for testing host susceptibility to the virus. While dogs appear susceptible to SARS-CoV-2, numerous reports have observed that MDCK cells are not permissive for virus replication, although at least one has reported low level replication (Barr et al., 2020; Hoffmann et al., 2020a; Wang et al., 2021). Previous research done in our laboratory and by others clearly demonstrate that poultry and other bird species cannot support replication of the virus (Schlottau et al., 2020; Shi et al., 2020; Suarez et al., 2020). We hypothesized that avian cell lines could become permissible to infection if they expressed a suitable ACE2 receptor and produced high enough levels of a protease that could activate SARS-CoV-2. It is worth mentioning that proteolytic cleavage of the S protein at the S1/S2 interface was assumed to be provided by furin-like enzymes naturally present in the DF1 or MDCK cell lines.
The SARS-CoV-2 utilizes the ACE2 protein as the primary receptor for entry into host cells and the TMPRSS2 protease has been shown to be critical for cleavage/activation of the spike protein (Gallagher and McCray, 2021; Sasaki et al., 2021). In these studies, the transgenic insertion of the human ACE2 and TMPRSS2 genes conferred virus attachment and replication ability in the non-permissive avian DF1 cell lines and MDCK cell lines. The results also demonstrated that single expression of either the human receptor or the protease was not sufficient to allow for virus replication in these cell lines, either through a lack of attachment or spike protein activation. The chicken ACE2 gene could be detected by RT-PCR, and it is likely not functional for attachment for SARS-CoV-2. These studies also demonstrate DF1 cells expressing the ACE2 and TMPRSS2 genes from different animal species can be used as an in vitro predictive model for virus replication. Wild type DF1 cells are normally incapable of supporting SARS-CoV-2 replication; however, expression of the receptor and protease genes from human, cat, goat and golden hamster allowed virus replication. This in vitro model correlates with the known natural or experimental susceptibility of three of these species and supports its use as a predictive model. The surprising result is the potential susceptibility of goats. Goats have not been known to be naturally or experimentally infected at this time, but one study has previously suggested that SARS-CoV-2 can infect HEK cells that are expressing goat ACE2 (Zhang et al., 2021). An experimental study in cattle did not show evidence of virus replication, and further work needs to be conducted to determine if goats are experimentally susceptible to infection.
Multiple studies have looked at experimental inoculation in pigs, swine cell lines, and in cell lines where the swine ACE2 gene has been expressed with mixed results. Three different experimental challenge studies with swine were conducted with 2 studies showing no infection and a third showing only a small number of pigs with any evidence of virus detection after challenge (Meekins et al., 2020; Pickering et al., 2021; Schlottau et al., 2020; Shi et al., 2020; Vergara-Alert et al., 2021). Two swine cell lines, swine testicular and porcine kidney cells, were also reported to develop CPE after several passages of virus. Multiple studies have also used different mammalian cell lines and transfected them with the swine ACE2 gene to allow for transient expression of the gene, and most found that SARS-CoV-2 or a SARS-CoV-2 pseudovirus could attach to and express protein in the cell as measured by several different methods (Conceicao et al., 2020; Li et al., 2019; Zhang et al., 2021; Zhao et al., 2020). This predictive data based on ACE2 data from some species, such as swine, suggest susceptibility to infection, although our results in DF1 cells did not show evidence of virus replication. One possible explanation for the discrepancy in animal studies and in vitro studies is that the ACE2 protein in swine is not efficiently expressed in the respiratory tract, which is the most likely route of exposure, and the virus cannot efficiently attach and infect the exposed pig (Zhai et al., 2020). Although results in swine are discordant, our studies using an avian cell line correlates closer with the swine challenge studies as we did not measure any virus in the cell supernatant that would be evidence of the virus completing the replication cycle, despite the possibility that the virus could attach to the modified cell line based on these previous studies.
The results with the horse ACE2 and TMPRSS2 genes showed no evidence of infection despite the relatively high sequence conservation of the horse ACE2 protein to human ACE2 at over 86%, which is higher than cats and Golden hamsters. Although the sequence similarity of human and horse ACE2 is high, the difficulty in challenging horses in a Biosafety level 3 animal facility has likely prevented the research from being performed. Our results provide additional support that horses are not susceptible to infection and do not need to be experimentally challenged.
Bats have been identified as likely reservoirs of both SARS-CoV-1 and MERS-CoV to humans through intermediate hosts including civet cats and dromedary camels, respectively (Corman et al., 2016; Ye et al., 2020; Zhou et al., 2021). The SARS-CoV-2 virus appears capable to bind to Little Brown bat and Great Roundleaf Bat ACE2 as observed by positive immunostaining and transient virus replication. However, the TMPRSS2 protease found in these species may not be functional as it lacks the 5’ terminus found in human and other animals, including other bat species. Analysis of the GenBank record suggests that only partial sequence is available and that the gene was not properly annotated and thus the gene sequences used in these studies may not represent the true open reading frame. Further research is required to determine whether the anomaly is a sequence artifact. However, at least one report suggests lower level binding with Little Brown bat ACE2 compared to human ACE2 with the SARS-Cov-2 virus due to zoonotic transmission from bats to humans (Conceicao et al., 2020). Our results also demonstrated lower levels of virus replication, although it is unclear if this is due to receptor binding or protease functionality. Further research is underway to determine the contribution of other bat species ACE2 and TMPRSS2 as a barrier to SARS-CoV-2 infection.
As noted, SARS-CoV-2 appears to have a broad host range among mammals, however the full host range is unknown. Predictive in silico studies based on ACE2 analysis have described potential broad host tropism of the virus to numerous species including cat, goat and hamster (Conceicao et al., 2020; Damas et al., 2020; Liu et al., 2021; Rendon-Marin et al., 2021). These studies also predict many aquatic species including whales and dolphins to have high likelihood of binding to SARS-CoV-2 spike protein. In silico analysis of the TMPRSS2 protein is less predictive, but the protease activation of the SARS-CoV-2 spike protein is necessary for replication to occur. As noted earlier, proteases other than TMPRSS2 have been demonstrated to have the ability to cleave the spike protein. In vivo testing of many large domestic animals and wild animal species would be difficult, if not impossible, because of the requirement for work to be performed in a secure biocontainment facility. Therefore, we propose this model could be utilized to screen many species for susceptibility to SARS-CoV-2 infection. Understanding the host range of SARS-CoV-2 is crucial to understanding the ecology of the virus and the role different species may play as reservoirs or bridge-species into humans. Species that can be infected also may be affected by disease. Our in vitro testing in DF1 ++ cells positively correlated with available in vivo challenge data. Taken together, the integration and expression of the ACE2 and TMPRRS2 from a target species in the otherwise non-permissive avian cell line provides a rapid and economical method to screen species for susceptibility to SARS-CoV-2.
CRediT authorship contribution statement
Darrell R. Kapczynski: Conceptualization, Methodology, Formal analysis, Writing – original draft, preparation, Writing – review & editing. Ryan Sweeney: Methodology, Formal analysis, Writing – original draft, preparation. Erica Spackman: Methodology, Formal analysis, Writing – original draft, preparation. Mary Pantin-Jackwood: Formal analysis. David L. Suarez: Conceptualization, Methodology, Formal analysis, Writing – original draft, preparation, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Linda Moon, Scott Lee, Suzanne DeBlois, and Tim Olivier for excellent technical assistance. This research was supported by funding from USDA, ARS, CRIS project #6040-32000-066-00D.
The following reagent was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate USA-WA1/2020, NR-52281. Vero African Green Monkey Kidney Cells (ATCC® CCL-81™), FR-243, was obtained through the International Reagent Resource, Influenza Division, WHO Collaborating Center for Surveillance, Epidemiology and Control of Influenza, Centers for Disease Control and Prevention, Atlanta, GA, USA.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virol.2022.01.014.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Barr I.G., Rynehart C., Whitney P., Druce J. SARS-CoV-2 does not replicate in embryonated hen's eggs or in MDCK cell lines. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.25.2001122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestle D., Heindl M.R., Limburg H., Van Lam van T., Pilgram O., Moulton H., Stein D.A., Hardes K., Eickmann M., Dolnik O., Rohde C., Klenk H.D., Garten W., Steinmetzer T., Bottcher-Friebertshauser E. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci Alliance. 2020;3 doi: 10.26508/lsa.202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brian D.A., Baric R.S. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- Conceicao C., Thakur N., Human S., Kelly J.T., Logan L., Bialy D., Bhat S., Stevenson-Leggett P., Zagrajek A.K., Hollinghurst P., Varga M., Tsirigoti C., Tully M., Chiu C., Moffat K., Silesian A.P., Hammond J.A., Maier H.J., Bickerton E., Shelton H., Dietrich I., Graham S.C., Bailey D. The SARS-CoV-2 Spike protein has a broad tropism for mammalian ACE2 proteins. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Eckerle I., Memish Z.A., Liljander A.M., Dijkman R., Jonsdottir H., Juma Ngeiywa K.J., Kamau E., Younan M., Al Masri M., Assiri A., Gluecks I., Musa B.E., Meyer B., Muller M.A., Hilali M., Bornstein S., Wernery U., Thiel V., Jores J., Drexler J.F., Drosten C. Link of a ubiquitous human coronavirus to dromedary camels. Proc. Natl. Acad. Sci. U. S. A. 2016;113:9864–9869. doi: 10.1073/pnas.1604472113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damas J., Hughes G.M., Keough K.C., Painter C.A., Persky N.S., Corbo M., Hiller M., Koepfli K.P., Pfenning A.R., Zhao H., Genereux D.P., Swofford R., Pollard K.S., Ryder O.A., Nweeia M.T., Lindblad-Toh K., Teeling E.C., Karlsson E.K., Lewin H.A. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc. Natl. Acad. Sci. U. S. A. 2020;117:22311–22322. doi: 10.1073/pnas.2010146117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Yue X., Xia H., Bindom S.M., Hickman P.J., Filipeanu C.M., Wu G., Lazartigues E. Angiotensin-converting enzyme 2 overexpression in the subfornical organ prevents the angiotensin II-mediated pressor and drinking responses and is associated with angiotensin II type 1 receptor downregulation. Circ. Res. 2008;102:729–736. doi: 10.1161/CIRCRESAHA.107.169110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni D., Cagliani R., Clerici M., Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher T., McCray P.B., Jr. The first few days of a SARS-CoV-2 infection viewed at single-cell resolution. PLoS Biol. 2021;19 doi: 10.1371/journal.pbio.3001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghai R.R., Carpenter A., Liew A.Y., Martin K.B., Herring M.K., Gerber S.I., Hall A.J., Sleeman J.M., VonDobschuetz S., Behravesh C.B. Animal reservoirs and hosts for emerging alphacoronaviruses and betacoronaviruses. Emerg. Infect. Dis. 2021;27:1015–1022. doi: 10.3201/eid2704.203945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacka I., Bertram S., Muller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K., Niemeyer D., Schneider H., Drosten C., Pohlmann S. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralinski L.E., Menachery V.D. 2020. Return of the Coronavirus: 2019-nCoV. Viruses 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H.J., Wen H.L., Zhou C.M., Chen F.F., Luo L.M., Liu J.W., Yu X.J. Bats as reservoirs of severe emerging infectious diseases. Virus Res. 2015;205:1–6. doi: 10.1016/j.virusres.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Duan X., Yang L., Nilsson-Payant B.E., Wang P., Duan F., Tang X., Yaron T.M., Zhang T., Uhl S., Bram Y., Richardson C., Zhu J., Zhao Z., Redmond D., Houghton S., Nguyen D.T., Xu D., Wang X., Jessurun J., Borczuk A., Huang Y., Johnson J.L., Liu Y., Xiang J., Wang H., Cantley L.C., tenOever B.R., Ho D.D., Pan F.C., Evans T., Chen H.J., Schwartz R.E., Chen S. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature. 2021;589:270–275. doi: 10.1038/s41586-020-2901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt J., Tamin A., Lu X., Kamili S., Sakthivel S.K., Murray J., Queen K., Tao Y., Paden C.R., Zhang J., Li Y., Uehara A., Wang H., Goldsmith C., Bullock H.A., Wang L., Whitaker B., Lynch B., Gautam R., Schindewolf C., Lokugamage K.G., Scharton D., Plante J.A., Mirchandani D., Widen S.G., Narayanan K., Makino S., Ksiazek T.G., Plante K.S., Weaver S.C., Lindstrom S., Tong S., Menachery V.D., Thornburg N.J. Severe acute respiratory syndrome coronavirus 2 from patient with coronavirus disease, United States. Emerg. Infect. Dis. 2020;26:1266–1273. doi: 10.3201/eid2606.200516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald-Sargent T., Gallagher T. Ready, set, fuse! the coronavirus spike protein and acquisition of fusion competence. Viruses. 2012;4:557–580. doi: 10.3390/v4040557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Hofmann-Winkler H., Smith J.C., Kruger N., Arora P., Sorensen L.K., Sogaard O.S., Hasselstrom J.B., Winkler M., Hempel T., Raich L., Olsson S., Danov O., Jonigk D., Yamazoe T., Yamatsuta K., Mizuno H., Ludwig S., Noe F., Kjolby M., Braun A., Sheltzer J.M., Pohlmann S. Camostat mesylate inhibits SARS-CoV-2 activation by TMPRSS2-related proteases and its metabolite GBPA exerts antiviral activity. EBioMedicine. 2021;65:103255. doi: 10.1016/j.ebiom.2021.103255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Pohlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78:779–784 e775. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280 e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulswit R.J., de Haan C.A., Bosch B.J. Coronavirus spike protein and tropism changes. Adv. Virus Res. 2016;96:29–57. doi: 10.1016/bs.aivir.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Yang H., Kapczynski D.R. Chicken interferon alpha pretreatment reduces virus replication of pandemic H1N1 and H5N9 avian influenza viruses in lung cell cultures from different avian species. Virol. J. 2011;8:447. doi: 10.1186/1743-422X-8-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapczynski D.R., Tumpey T.M., Hidajat R., Zsak A., Chrzastek K., Tretyakova I., Pushko P. Vaccination with virus-like particles containing H5 antigens from three H5N1 clades protects chickens from H5N1 and H5N8 influenza viruses. Vaccine. 2016;34:1575–1581. doi: 10.1016/j.vaccine.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.M., Kim H.M., Lee E.J., Jo H.J., Yoon Y., Lee N.J., Son J., Lee Y.J., Kim M.S., Lee Y.P., Chae S.J., Park K.R., Cho S.R., Park S., Kim S.J., Wang E., Woo S., Lim A., Park S.J., Jang J., Chung Y.S., Chin B.S., Lee J.S., Lim D., Han M.G., Yoo C.K. Detection and isolation of SARS-CoV-2 in Serum, urine, and stool specimens of COVID-19 patients from the Republic of Korea. Osong Public Health Res Perspect. 2020;11:112–117. doi: 10.24171/j.phrp.2020.11.3.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J., Group S.W. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B.G., Paul van Schayck J., Mykytyn A.Z., Duimel H.Q., van Donselaar E., Riesebosch S., Kuijpers H.J.H., Schipper D., van de Wetering W.J., de Graaf M., Koopmans M., Cuppen E., Peters P.J., Haagmans B.L., Clevers H. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Tomlinson A.C., Wong A.H., Zhou D., Desforges M., Talbot P.J., Benlekbir S., Rubinstein J.L., Rini J.M. The human coronavirus HCoV-229E S-protein structure and receptor binding. Elife. 2019;8 doi: 10.7554/eLife.51230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.Y., Li Y.T., Chen Y.T., Chen T.C., Hu C.J., Chen H.W. Identification of an infectious bronchitis coronavirus strain exhibiting a classical genotype but altered antigenicity, pathogenicity, and innate immunity profile. Sci. Rep. 2016;6:37725. doi: 10.1038/srep37725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Hu G., Wang Y., Ren W., Zhao X., Ji F., Zhu Y., Feng F., Gong M., Ju X., Zhu Y., Cai X., Lan J., Guo J., Xie M., Dong L., Zhu Z., Na J., Wu J., Lan X., Xie Y., Wang X., Yuan Z., Zhang R., Ding Q. vol. 118. Proc Natl Acad Sci U S A; 2021. (Functional and Genetic Analysis of Viral Receptor ACE2 Orthologs Reveals a Broad Potential Host Range of SARS-CoV-2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Zheng H., Lin H., Li M., Yuan R., Peng J., Xiong Q., Sun J., Li B., Wu J., Yi L., Peng X., Zhang H., Zhang W., Hulswit R.J.G., Loman N., Rambaut A., Ke C., Bowden T.A., Pybus O.G., Lu J. Identification of common deletions in the spike protein of severe acute respiratory syndrome coronavirus 2. J. Virol. 2020;94 doi: 10.1128/JVI.00790-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Wang L., Sakthivel S.K., Whitaker B., Murray J., Kamili S., Lynch B., Malapati L., Burke S.A., Harcourt J., Tamin A., Thornburg N.J., Villanueva J.M., Lindstrom S. US CDC real-time Reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020;26 doi: 10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meekins D.A., Morozov I., Trujillo J.D., Gaudreault N.N., Bold D., Carossino M., Artiaga B.L., Indran S.V., Kwon T., Balaraman V., Madden D.W., Feldmann H., Henningson J., Ma W., Balasuriya U.B.R., Richt J.A. Susceptibility of swine cells and domestic pigs to SARS-CoV-2. Emerg. Microb. Infect. 2020;9:2278–2288. doi: 10.1080/22221751.2020.1831405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteil V., Kwon H., Prado P., Hagelkruys A., Wimmer R.A., Stahl M., Leopoldi A., Garreta E., Hurtado Del Pozo C., Prosper F., Romero J.P., Wirnsberger G., Zhang H., Slutsky A.S., Conder R., Montserrat N., Mirazimi A., Penninger J.M. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913 e907. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M.A., Corman V.M., Jores J., Meyer B., Younan M., Liljander A., Bosch B.J., Lattwein E., Hilali M., Musa B.E., Bornstein S., Drosten C. MERS coronavirus neutralizing antibodies in camels, Eastern Africa, 1983-1997. Emerg. Infect. Dis. 2014;20:2093–2095. doi: 10.3201/eid2012.141026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel V.B., Zhong J.C., Grant M.B., Oudit G.Y. Role of the ACE2/angiotensin 1-7 Axis of the renin-angiotensin system in heart failure. Circ. Res. 2016;118:1313–1326. doi: 10.1161/CIRCRESAHA.116.307708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering B.S., Smith G., Pinette M.M., Embury-Hyatt C., Moffat E., Marszal P., Lewis C.E. Susceptibility of domestic swine to experimental infection with severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2021;27:104–112. doi: 10.3201/eid2701.203399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendon-Marin S., Martinez-Gutierrez M., Whittaker G.R., Jaimes J.A., Ruiz-Saenz J. SARS CoV-2 spike protein in silico interaction with ACE2 receptors from wild and domestic species. Front. Genet. 2021;12:571707. doi: 10.3389/fgene.2021.571707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M., Uemura K., Sato A., Toba S., Sanaki T., Maenaka K., Hall W.W., Orba Y., Sawa H. SARS-CoV-2 variants with mutations at the S1/S2 cleavage site are generated in vitro during propagation in TMPRSS2-deficient cells. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlottau K., Rissmann M., Graaf A., Schon J., Sehl J., Wylezich C., Hoper D., Mettenleiter T.C., Balkema-Buschmann A., Harder T., Grund C., Hoffmann D., Breithaupt A., Beer M. SARS-CoV-2 in fruit bats, ferrets, pigs, and chickens: an experimental transmission study. Lancet Microbe. 2020;1:e218–e225. doi: 10.1016/S2666-5247(20)30089-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B., Liu R., He X., Shuai L., Sun Z., Zhao Y., Liu P., Liang L., Cui P., Wang J., Zhang X., Guan Y., Tan W., Wu G., Chen H., Bu Z. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulla A., Heald-Sargent T., Subramanya G., Zhao J., Perlman S., Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 2011;85:873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H.D., Tu C.C., Zhang G.W., Wang S.Y., Zheng K., Lei L.C., Chen Q.X., Gao Y.W., Zhou H.Q., Xiang H., Zheng H.J., Chern S.W., Cheng F., Pan C.M., Xuan H., Chen S.J., Luo H.M., Zhou D.H., Liu Y.F., He J.F., Qin P.Z., Li L.H., Ren Y.Q., Liang W.J., Yu Y.D., Anderson L., Wang M., Xu R.H., Wu X.W., Zheng H.Y., Chen J.D., Liang G., Gao Y., Liao M., Fang L., Jiang L.Y., Li H., Chen F., Di B., He L.J., Lin J.Y., Tong S., Kong X., Du L., Hao P., Tang H., Bernini A., Yu X.J., Spiga O., Guo Z.M., Pan H.Y., He W.Z., Manuguerra J.C., Fontanet A., Danchin A., Niccolai N., Li Y.X., Wu C.I., Zhao G.P. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. U. S. A. 2005;102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez D.L., Pantin-Jackwood M.J., Swayne D.E., Lee S.A., DeBlois S.M., Spackman E. Lack of susceptibility to SARS-CoV-2 and MERS-CoV in poultry. Emerg. Infect. Dis. 2020;26:3074–3076. doi: 10.3201/eid2612.202989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y.J., Nikolaienko S.I., Dibrova V.A., Dibrova Y.V., Vasylyk V.M., Novikov M.Y., Shults N.V., Gychka S.G. SARS-CoV-2 spike protein-mediated cell signaling in lung vascular cells. Vasc. Pharmacol. 2021;137:106823. doi: 10.1016/j.vph.2020.106823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara-Alert J., Rodon J., Carrillo J., Te N., Izquierdo-Useros N., Rodriguez de la Concepcion M.L., Avila-Nieto C., Guallar V., Valencia A., Cantero G., Blanco J., Clotet B., Bensaid A., Segales J. Pigs are not susceptible to SARS-CoV-2 infection but are a model for viral immunogenicity studies. Transbound Emerg Dis. 2021;68(4):1721–1725. doi: 10.1111/tbed.13861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Fan X., Bonenfant G., Cui D., Hossain J., Jiang N., Larson G., Currier M., Liddell J., Wilson M., Tamin A., Harcourt J., Ciomperlik-Patton J., Pang H., Dybdahl-Sissoko N., Campagnoli R., Shi P.Y., Barnes J., Thornburg N.J., Wentworth D.E., Zhou B. Susceptibility to SARS-CoV-2 of cell lines and substrates commonly used to diagnose and isolate influenza and other viruses. Emerg. Infect. Dis. 2021;27:1380–1392. doi: 10.3201/eid2705.210023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z.W., Yuan S., Yuen K.S., Fung S.Y., Chan C.P., Jin D.Y. Zoonotic origins of human coronaviruses. Int. J. Biol. Sci. 2020;16:1686–1697. doi: 10.7150/ijbs.45472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang R., Gomez Castro M.F., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., Liu Z., Brulois K.F., Wang X., Greenberg H.B., Diamond M.S., Ciorba M.A., Whelan S.P.J., Ding S. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai X., Sun J., Yan Z., Zhang J., Zhao J., Zhao Z., Gao Q., He W.T., Veit M., Su S. Comparison of severe acute respiratory syndrome coronavirus 2 spike protein binding to ACE2 receptors from human, pets, farm animals, and putative intermediate hosts. J. Virol. 2020;94 doi: 10.1128/JVI.00831-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.L., Li Y.M., Sun J., Zhang Y.Y., Wang T.Y., Sun M.X., Wang M.H., Yang Y.L., Hu X.L., Tang Y.D., Zhao J., Cai X. Evaluating angiotensin-converting enzyme 2-mediated SARS-CoV-2 entry across species. J. Biol. Chem. 2021;296:100435. doi: 10.1016/j.jbc.2021.100435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., To K.K.W., Sze K.H., Yung T.T., Bian M., Lam H., Yeung M.L., Li C., Chu H., Yuen K.Y. A broad-spectrum virus- and host-targeting peptide against respiratory viruses including influenza virus and SARS-CoV-2. Nat. Commun. 2020;11:4252. doi: 10.1038/s41467-020-17986-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Li C., Liu X., Chiu M.C., Zhao X., Wang D., Wei Y., Lee A., Zhang A.J., Chu H., Cai J.P., Yip C.C., Chan I.H., Wong K.K., Tsang O.T., Chan K.H., Chan J.F., To K.K., Chen H., Yuen K.Y. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med. 2020;26:1077–1083. doi: 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]
- Zhou P., Fan H., Lan T., Yang X.L., Shi W.F., Zhang W., Zhu Y., Zhang Y.W., Xie Q.M., Mani S., Zheng X.S., Li B., Li J.M., Guo H., Pei G.Q., An X.P., Chen J.W., Zhou L., Mai K.J., Wu Z.X., Li D., Anderson D.E., Zhang L.B., Li S.Y., Mi Z.Q., He T.T., Cong F., Guo P.J., Huang R., Luo Y., Liu X.L., Chen J., Huang Y., Sun Q., Zhang X.L., Wang Y.Y., Xing S.Z., Chen Y.S., Sun Y., Li J., Daszak P., Wang L.F., Shi Z.L., Tong Y.G., Ma J.Y. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556:255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Hui K.P.Y., So R.T.Y., Lv H., Perera R., Chu D.K.W., Gelaye E., Oyas H., Njagi O., Abayneh T., Kuria W., Walelign E., Wanglia R., El Masry I., Von Dobschuetz S., Kalpravidh W., Chevalier V., Miguel E., Fassi-Fihri O., Trarore A., Liang W., Wang Y., Nicholls J.M., Zhao J., Chan M.C.W., Poon L.L.M., Mok C.K.P., Peiris M. Phenotypic and genetic characterization of MERS coronaviruses from Africa to understand their zoonotic potential. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2103984118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmora P., Hoffmann M., Kollmus H., Moldenhauer A.S., Danov O., Braun A., Winkler M., Schughart K., Pohlmann S. TMPRSS11A activates the influenza A virus hemagglutinin and the MERS coronavirus spike protein and is insensitive against blockade by HAI-1. J. Biol. Chem. 2018;293:13863–13873. doi: 10.1074/jbc.RA118.001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.