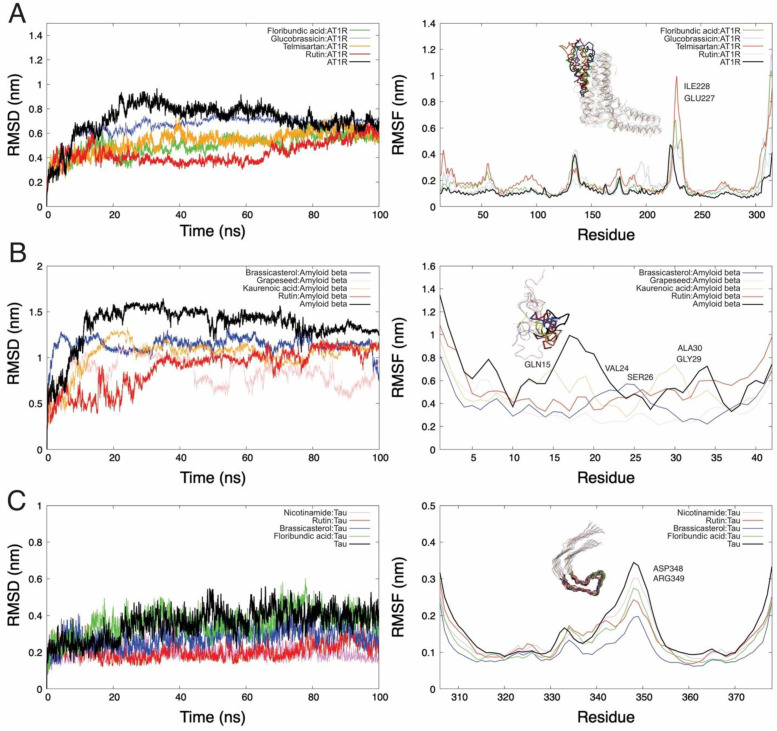
Figure 2.
Representation of DM trajectories over 100 ns for AT1R, amyloid beta, and tau. The figures on the right side are the RMSD plots, and those on the left side are the RMSF plots of the backbone per residue. (A) The tendency of the RMSD plot for the AT1R–rutin system shows that the protein does not constantly deviate from its original conformation even though this compound generates the largest fluctuations in AT1R (RMSF analysis); (B) the RMSD plot for amyloid beta in its native state (black line) shows a high structural instability due to the size and lack of other secondary conformations. However, the coupled systems reduce this deviation; (C) tau with ligands shows a tendency of high conformational stability.