Table 1.
Representative cell-permeable MPs reported since 2017. CPP motifs are highlighted in green color.
| Compd No. |
Compound structure | Target (KD
or IC50 value for target) |
Biological activity |
Cell entry mechanism |
Refs |
|---|---|---|---|---|---|
| 1 |
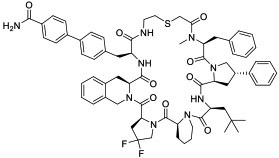
|
NNMT(IC50 = 1.9 nM for human NNMT) | In vitro (IC50 = 0.77 μM for NNMT inhibition in HEK293 cells) | Passive diffusion | [25] |
| 2 |
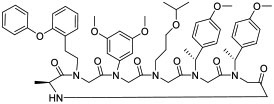
|
β-catenin (IC50 = 5.4 μM) | In vitro; in vivo (rescued eye development in zebrafish model) | Passive diffusion | [26] |
| 3 |
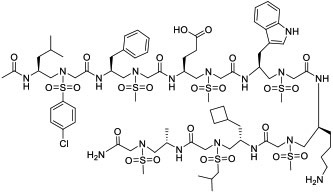
|
MDM2 (KD = 26 nM) | In vitro by stapled analog (activated p53 in U2OS cells at 30 μM) | Potentially passive diffusion | [28] |
| 4 |
|
TEAD (KD = 0.7 pM for human TEAD) | In vitro (activated Hippo-associated genes in cardiomyocytes at 30 μM) | Endocytosis | [33] |
| 5 |
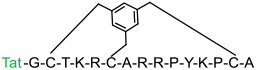
|
RbAp48 (KD = 8.6 nM) | In vitro (increased p53 level in U2OS cells) | Endocytosis | [34] |
| 6 |
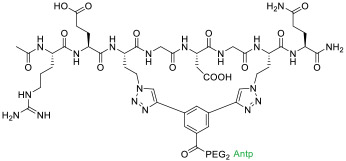
|
Tankyrase (KD = 440 nM) | In vitro (decreased Wnt reporter activity in HEK293T cells [IC50 ≈ 50 μM]) | Endocytosis | [36] |
| 7 |

|
Jun subunit of transcription factor activator protein-1 (KD =2 μM) | In vitro (antiproliferative activity against MCF-7 and ZR75-1 breast cancer cells) | Endocytosis | [37] |
| 8 |

|
HDAC (IC50 = 99-303 nM for different HDAC isoforms) | In vitro; in vivo (TGI @ 50 mg/kg in PA-1 and NTERA-2 mouse xenograft models) | Endocytosis | [38] |
| 9 |
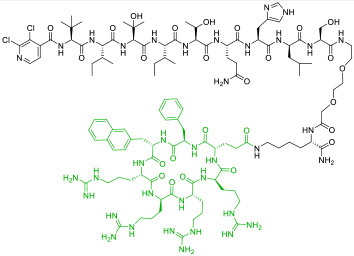
|
Calcineurin (KD = 16 nM) | In vitro; in vivo (prevented acute lung injury @ 3-5 mg/kg in a mouse model) | Endocytosis | [41] |
| 10 |
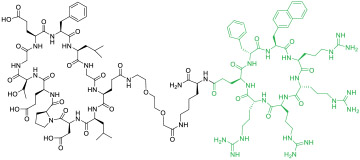
|
Keapl (IC50 = 153 nM) | In vitro (inhibited Keap1/Nrf2 PPI in ARE reporter-HepG2 cell line [EC50 ≈ 1.1 μM]) | Endocytosis | [43] |
| 11 |

|
Pin1 (IC50 = 220 nM) | In vitro (inhibited the proliferation of HeLa cervical cancer cells) | Endocytosis | [46] |
| 12 |
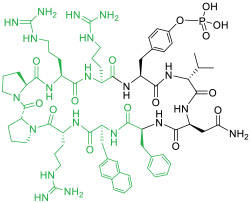
|
Grb2 SH2 domain (IC50 = 400 nM) | In vitro (inhibited Grb2/Ras signaling in MDA-MB-468 breast cancer cells [IC50 ≈ 15 MM]) | Endocytosis | [47] |
| 13 |
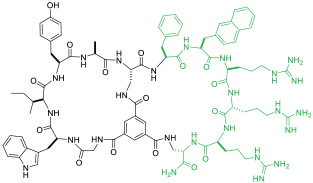
|
NEMO(IC50 = 1.0 μM) | In vitro (inhibition of TNFα-induced NF-κB activation [IC50 = 10 μM]) | Endocytosis | [49] |
| 14 |
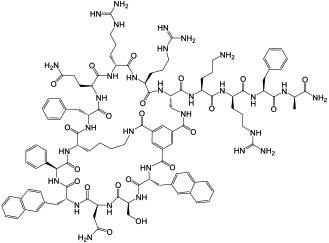
|
K-, H-, and N-Ras (KD = 21 nM for KRasG12V-GppNHp) | In vitro; in vivo (TGI @ 1-5 mg/kg in A549 and H358 mouse xenograft models) | Endocytosis | [50] |
| 15 |
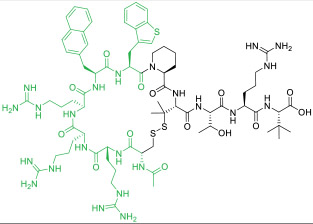
|
PDZ domain of CFTR-associated ligand (KD = 6 nM) | In vitro; ex vivo (improved ion channel activity of F508del-CFTR by 3-fold [EC50 ≈ 10 nM]) | Endocytosis | [51] |
| 16 |
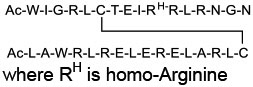
|
Ras (KD = 2 μM for HRas) | In vitro (antiproliferative activity in Ras mutant cancer cells) | Endocytosis | [53] |
| 17 |
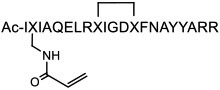
|
Bcl2A1 (apparent Ki <0.1 nM) | In vitro (covalent modification of Bcl2A1 in U937 lymphoma cells) | Unknown | [68] |
| 18 |

|
β-catenin (KD = 123 nM) | In vitro (antiproliferative effect in Wnt-addicted colorectal cancer cells DLD-1 and SW480) | Unknown | [69] |
| 19 |
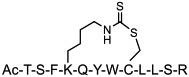
|
MDM2/X (KD = 0.87 and 3.9 nM for MDM2 and MDMX) | In vitro (antiproliferative effect in HCT116 cells [IC50 ≈ 25 μM]) | Unknown | [71] |
| 20 |
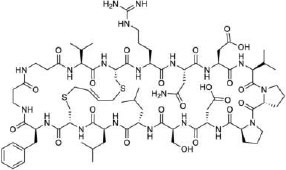
|
β-catenin (KD = 91 nM) | In vitro (inhibition of Wnt signaling in HEK293T reporter cell line [IC50 = 8 μM]) | Unknown | [74] |
| 21 |
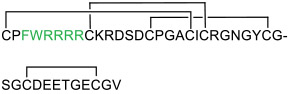
|
Keap1 (KD = 35 nM) | In vitro (increased the expression of Nrf2-regulated genes in HEK293T cells) | Unknown (potentially endocytosis) | [79] |
| 22 |
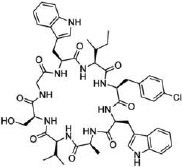
|
UEV domain of TSG101 protein (KD = 11.9 μM) | In vitro (viruslike particle budding assay in HEK293T cells [IC50 ≈ 2 μM]) | Unknown | [83] |
| 23 |
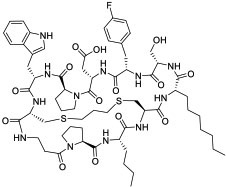
|
KRasG12D (EC50 = 22 nM) | In vitro; in vivo (TGI @40 mg/kg in PANC-1 mouse xenograft model) | Unknown | [84] |
| 24 |
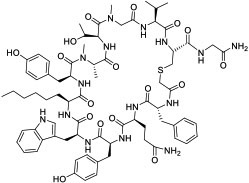
|
K48Ub tetramer (KD = 9 nM) | In vitro; in vivo (TGI @1 mg/kg in mouse model) | Unknown | [85] |
| 25 |
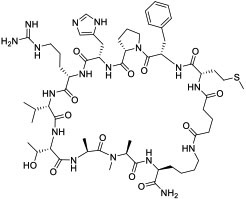
|
LC (KD = 120 and 192 nM for LC3A and LC3B, respectively) | In vitro (inhibited autophagy in HeLa cervical cancer cells) | Unknown | [87] |
| 26 |
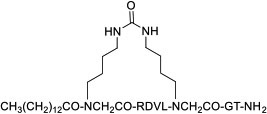
|
MyD88 | In vivo (reduced disease severity @ 10 mg/kg in sclerosis mouse models) | Unknown | [88] |
