Abstract
Artificial intelligence (AI) is transforming many domains, including finance, agriculture, defense, and biomedicine. In this paper, we focus on the role of AI in clinical and translational research (CTR), including preclinical research (T1), clinical research (T2), clinical implementation (T3), and public (or population) health (T4). Given the rapid evolution of AI in CTR, we present three complementary perspectives: (1) scoping literature review, (2) survey, and (3) analysis of federally funded projects. For each CTR phase, we addressed challenges, successes, failures, and opportunities for AI. We surveyed Clinical and Translational Science Award (CTSA) hubs regarding AI projects at their institutions. Nineteen of 63 CTSA hubs (30%) responded to the survey. The most common funding source (48.5%) was the federal government. The most common translational phase was T2 (clinical research, 40.2%). Clinicians were the intended users in 44.6% of projects and researchers in 32.3% of projects. The most common computational approaches were supervised machine learning (38.6%) and deep learning (34.2%). The number of projects steadily increased from 2012 to 2020. Finally, we analyzed 2604 AI projects at CTSA hubs using the National Institutes of Health Research Portfolio Online Reporting Tools (RePORTER) database for 2011–2019. We mapped available abstracts to medical subject headings and found that nervous system (16.3%) and mental disorders (16.2) were the most common topics addressed. From a computational perspective, big data (32.3%) and deep learning (30.0%) were most common. This work represents a snapshot in time of the role of AI in the CTSA program.
Keywords: artificial intelligence, machine learning, translational medical research
INTRODUCTION AND BACKGROUND
Artificial intelligence (AI) has experienced multiple “boom and bust” cycles since the term was first coined by computer and cognitive scientist John McCarthy in 1955. Historically biomedical AI applications (Figure 1) included symbolic systems (e.g., HELP system 1 and subsequent Arden Syntax, 2 MYCIN, 3 and INTERNIST‐I 4 ) and statistical or “subsymbolic” (e.g., Leeds abdominal pain system 5 ).
FIGURE 1.
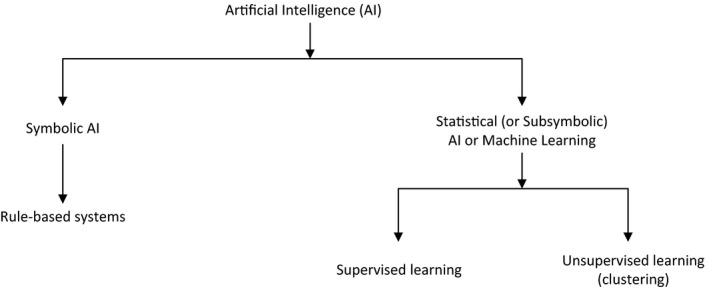
A Artificial intelligence (AI) concepts
Perhaps the most popular symbolic approaches were rule‐based systems (including the Arden syntax and MYCIN) that implemented “if‐then” rules. For example, consider the following MYCIN rule 3 :
IF: 1) The gram stain of the organism is gram neg, and
2) The morphology of the organism is rod, and
3) The aerobicity of the organism is anaerobic
THEN: There is suggestive evidence (0.6) that the identity of the organism is bacteroides.
Rule‐based approaches are intuitively easy to understand, but may not work well for complex applications typical in biomedicine. Large collections of rules (“rule bases”) can be difficult to maintain and explicitly encoding the many exceptions that are common in biomedicine (e.g., oral temperature may not be reliable if the patient just drank a hot beverage) is impractical. 6
Most recent approaches are statistical and are often referred to as “machine learning” (ML). Many statistical systems implement some version of supervised ML where the algorithm requires a “training set” of labeled examples (e.g., patients with vs. without a particular disease). The algorithm learns to differentiate between positive and negative examples based on “features” (e.g., laboratory test results and clinical findings) that are associated with the outcome of interest (e.g., presence of the disease). In contrast, unsupervised ML algorithms cluster similar cases together without the need for a labeled training set. Thus, an unsupervised algorithm can report that “these cases are similar to each other,” but not what the similarity means.
More recently, a particular class of ML models called “deep learning” has become popular due to the models’ ability to solve problems that cannot be defined precisely, such as differentiating malignant from benign skin lesions based on images of the lesions. 7 Deep learning models rely on multilayer (thus “deep”) neural networks. Although neural networks were described over 60 years ago, 8 they became much more useful with modern computers applied to very large data sets that were not previously available. We are currently in an “AI boom” phase, fueled by increased availability of large clinical and research data sets (“big data”), rapid development of novel statistical algorithms that leverage big data, and the ubiquitous access to faster, cheaper, and ever more powerful computers.
At least two communities are involved in biomedical AI. The first group is comprised of “methodologists,” including statisticians, machine learning experts, and computer and “data” scientists. The second community is comprised of “domain experts,” including biomedical scientists, clinicians, and healthcare administrators who understand the problems and the data but too often are working separately from the methodologists. These communities have very different perspectives, including largely separate literatures, conferences, academic promotion criteria, and cultures. A particular challenge is to identify specific important problems that can be addressed by AI given available data and algorithms where the results can meaningfully impact clinical care. 9 This may be one reason why multiple research and clinical applications have been developed, but relatively few applications have successfully transitioned from algorithms that perform well on standardized data sets to operational systems demonstrated to improve clinical outcomes.
Since 2006, the CTSA program has included efforts to bring methodologists and domain experts together in service of translational science and clinical application. CTSA institutions or “hubs” have explicitly been committed to clinical and translational science and each CTSA hub includes defined efforts in biomedical informatics as well as biostatistics. These efforts ideally include both methodologists and domain experts. Thus, the CTSA hubs are likely to be leaders in the application of AI to clinical and translational science.
Motivated by the desire to facilitate clinically relevant applications of AI, we present three complementary “views” onto this rapidly evolving field. First, we conducted a scoping review 10 of biomedical AI efforts in the published literature. We adopted the National Center for Advancing Translational Science (NCATS) vision of translational science, 11 to identify challenges and opportunities for AI across the translational science spectrum. However, the published literature provides an incomplete view. For example, applications implemented and maintained by the clinical enterprise may not be described in publications. Thus, we surveyed CTSA hubs to self‐identify existing, funded AI projects at CTSA hubs. Finally, to identify additional projects and ongoing work, we analyzed biomedical AI projects at CTSA hubs funded by the US National Institutes of Health (NIH) and the US National Science Foundation (NSF).
SCOPING REVIEW METHODS
The search strategy was created by the reference team at the Houston Academy of Medicine–Texas Medical Center (HAM‐TMC) library. Customized search queries were created for each phase of clinical and translational science (T1–4). Searches were executed during the week of August 10, 2020, using Ovid (https://www.ovid.com) that includes MEDLINE‐indexed literature as well as additional PubMed articles. Ovid was available via the HAM‐TMC library. Supplementary Material SA describes the precise queries.
Inclusion criteria were difficult to define precisely. For example, how to determine whether a publication described a quality improvement project that was later published or a clinical application (i.e., T2 or T3 research). Thus, the first author reviewed all titles/abstracts to categorize references as “not relevant,” “possibly relevant,” or “relevant” (see table in Supplementary Material SA). In addition, the results for each section were independently reviewed by a second author for inclusion. Section authors were also free to add references outside of the systematic search strategy. The section authors made the final decision to include or exclude a particular reference.
PRECLINICAL RESEARCH (T1)
Challenges
Early drug development is a prototypical and challenging preclinical research area. Important areas in early drug development include methods to interpret high throughput data for target discovery or target selection, in silico modeling for drug discovery, prioritizing potential compounds for synthesis, and experimental validation. Some of the methods and databases in this space have recently been reviewed by Rifaioglu et al. 12 Once potential drug candidates are synthesized, there is a pressing need to increase efficiency and speed of drug screening as well as to optimize the most promising compound (the lead compound) that has activity against the target, to increase efficacy, decrease toxicity, and achieve optimum pharmacokinetics (i.e., lead optimization). Specific challenges include: (1) rapidly testing combination therapies with the lead compounds early in drug development, (2) identifying new targets for existing compounds for drug repurposing, (3) model pharmacokinetics and pharmacodynamics to predict optimal dose, (4) identify and validate relevant biomarkers, and (5) assess safety and efficacy during early in vivo studies.
There are several reports of predicting drug response using ML or deep neural networks, often integrating molecular features. 13 , 14 , 15 Chen and Zhang did a recent survey and systematic assessment of four classical methods and 13 computational methods for drug response prediction using four public drug/response datasets. They hypothesized that drug response prediction can be framed as a supervised learning problem, and that given a training drug/response dataset with cell lines and drugs, a response can be learned and subsequently used to predict the response to: (1) a known drug (in the training dataset) in a new cell line, (2) a new drug in a known cell line, or (3) a new drug in a new cell line. The performance of different classes of computational models varied, but the overall performance suggested that there is still much room for improvement. They also found with existing methods, mutation, copy number variants, and methylation profiles of cell lines and drug grouping contributed little to drug response prediction when added to the gene expression profile. How to best integrate omics data remains an open question. In addition, we know that each cell line is unique and AI approaches that can “personalize” treatments to individual molecular profiles may be useful.
Successes
Processing and interpreting data from high‐throughput studies is an important area. For example, proteomics based on liquid chromatography with tandem mass spectrometry (LC‐MS/MS) is a well‐established approach for biomarker discovery and target validation. This technology generates massive data sets that are almost impossible to interpret manually. Zohora et al. proposed a deep learning‐based model, Deepiso, that combines convolutional and recurrent neural networks to detect peptide features of different charge states, as well as to estimate their intensity. 16 They demonstrated that the peptide feature list reported by Deepiso matched with 97.43% of high‐quality MS/MS identifications in a benchmark dataset, which was higher than several widely used tools. The model also could be “retrained” with missed features to evolve and improve.
Failures
There is a growing body of literature describing AI applications for target or drug discovery, to identify biomarkers of response or to select therapy. Unfortunately, many preclinical papers have predictions with no or limited validation. Recent applications of deep learning and AI for in silico drug discovery are promising. However, due in part to the long time required to develop a new drug, few such findings have entered clinical trials. Some targets identified using virtual screening have already failed to achieve clinical success. 17 Thus overall, AI has not yet delivered on its promise as many AI studies have not been translated into approved drugs, biomarkers, or clinical therapy selection algorithms.
Opportunities
Although drug discovery using AI is still in its infancy, this area still has much potential, and many pharmaceutical companies have started to invest in AI strategies for discovery. Gene function and gene variant annotation, biomarker discovery, and literature mining, are all examples of areas where data are being generated too rapidly for manual approaches and AI‐based tools will be essential for progress. With evolution of new fields, there may be new needs emerging that can leverage AI, such as immuno‐oncology and the need to predict tumor neoantigens. 18 In translational research, there is growing interest in simultaneously assessing multiple markers, such as use of multiplex immunohistochemistry in oncology, 19 , 20 assessing features of the tumor as well as the microenvironment. 21 Deep learning‐based image analysis is also being explored. 22
Application of AI to basic and translational research is a clear opportunity for team science. It is essential that basic and translational researchers learn more about the capabilities of AI. There are many AI‐derived predictions of new drugs, new indications for existing drugs, or new biomarkers. Commercial (start‐up) companies that leverage AI to improve drug discovery have an important role to play in realizing the potential of AI in preclinical research, sometimes cooperating with academic institutions resulting in research alliances. Multidisciplinary teams consisting of computer scientists, bioinformaticians, clinical informaticians, basic and translational researchers, and clinical domain experts can ensure the clinical potential of these findings can be assessed, with promising findings moving through preclinical and into clinical testing.
CLINICAL RESEARCH (T2)
Challenges
Perhaps the most important challenge to application of AI in clinical research may be the clinical environment itself. The rising complexity and cost of development, particularly developing and testing new therapies, is a major challenge for clinical research. 23 Specifically, challenges related to eligibility screening, data collection, and data verification have consistently been reported 23 , 24 , 25 , 26 , 27 as has failure of well‐controlled clinical trials to generalize to real‐world practice. 28 These challenges continue to urge consideration of observational study designs that leverage real‐world evidence. 29 Over the last decade, informatics approaches, such as AI, have been called to help address these challenges. 23 , 24
Another challenge is the significant human effort required to construct large training data sets required for accurate supervised ML. As an example, a system that achieved dermatologist‐level classification of skin lesions was trained on 129,450 labeled images. 7 Further, even when training data are available, human expertise is required to implement the model in a particular environment. Thus, significant effort has been devoted to decrease the human effort and expertise required for ML, including algorithms to automate feature selection and optimize hyperparameters (i.e., parameters required by the ML model). A recent review identified limitations of existing ML approaches in healthcare, such as inability to consistently perform across the size and variety of data within biomedicine, and limited demonstration in health care. 30
Successes
Of the 100 distinct articles identified in our search, 39 described development or application of AI in clinical research. An additional 23 described clinical uses of AI, such as outcome prediction or classification that could also be applied in clinical studies. Promising approaches leveraging the complementary strengths of humans and computers have been developed. 31 These approaches recognize that computers can support, rather than replace, human decision making. For example, relevant data can be selected using AI and presented to a human who can use a resulting visualization to make better decisions. Signal processing (e.g., electroencephalography 32 ), electronic health record (EHR) phenotyping (i.e., identifying patients with specific conditions or characteristics, such as smoking status 33 ), various predictive models (e.g., sepsis 34 and breast cancer progression 35 ) have been developed, validated, and found to perform well on the validation data set.
Failures
Clinical research poses multiple challenges to ML. For example, computer programs used to process data submitted for regulatory decision making require validation, as described in the code of federal regulations. 36 If the requirements in the US Food and Drug Administration (FDA) guidance on patient‐reported outcomes 37 were applied to AI, developers would be required to demonstrate validity or comparable performance in the data set and the patient population under study. The potential for bias due to under‐representation of groups during model development is well‐known. In other words, ML models are limited by the data used for development. These data may differ in unexpected ways from additional data in which they will be applied. Over‐ or under‐fitting has the potential to cause inaccuracy or bias in study data. Operation at scale is also particularly challenging in clinical research settings. For example, the variety of data sources used in clinical studies has significantly increased the challenge of achieving and verifying performance in new data sources and new data sets. 38 , 39 Further, the data used in a clinical study are specific to the research question. As such, information systems used for clinical studies require some level of customization to support the study, including specific data elements, data flow, or workflow. Study‐to‐study variation poses challenges to the use of general AI pipelines. Simultaneously, time pressures require application of existing or at least easily configurable solutions.
Opportunities
Resource constrained academic settings favor grant‐funded AI development and demonstration rather than AI infrastructure implemented at scale. Further, demonstration of new technology in industry funded clinical studies have historically been less rigorous and published in the trade literature, or viewed as trade secrets and infrequently published. 40 A recent industry meeting on AI in clinical research hosted by the European Medicines Agency featured presentation of multiple opportunities for application of AI in clinical research, including clinical data management—such as data cleaning, fraud detection, protocol violation trend detection, building and validation of synthetic control arms, and classification of clinical events, conditions, or findings. Demonstration projects for each were presented, only one of which was published in the peer‐reviewed literature. 41 , 42
There is an opportunity to identify “digital biomarkers” based on algorithms processing various data sets, perhaps derived from wearable technology, such as smart watches, smart phones, or pedometers. 43 For example, output of wearables, such as pedometers that count steps, could be useful to monitor vulnerable elderly patients at risk for rapid deterioration. However, AI may be useful to distinguish variation due to new or worsening illness from other causes (e.g., fewer steps while travelling).
There are multiple opportunities for AI to facilitate clinical research processes. For example, AI has the potential to make previously subjective assessment more objective (e.g., assessment of conjunctival hyperemia 44 ) and automatically identify potentially eligible patients for clinical trials. 45
CLINICAL IMPLEMENTATION (T3)
Challenges
A fundamental challenge is to translate the many promising AI advances that could potentially transform healthcare into clinical practice. Clinical implementation requires validation to assess algorithm performance in “real‐world” clinical settings. However, these are often lacking. 46 These studies are important to ensure patient safety and reliability by building trust and accountability. 47 , 48 Ultimately, healthcare providers and patients must both accept and trust the recommendations provided by the system. 48
Another challenge is the human‐computer interactions, which ensure that the technology is user‐friendly with interpretable and actionable output. 31 Additional challenges include the inability of existing EHR workflows to integrate the technology as well as legal and ethical issues, especially those relating to accountability and privacy. 47 , 48 These challenges must be overcome and often require an interdisciplinary team in all phases of implementation. 49
Successes
Successful clinical implementation of AI technology into existing workflows include the machine learning model, Sepsis Watch, occurring in three phases over 2 years at Duke Health. 47 Another example is the IDx‐DR device used in patients with diabetes to detect greater than mild diabetic retinopathy and macular edema. After completing a successful clinical trial with 900 patients, the FDA approved IDx‐DR in 2018 making it the first commercially available, autonomous AI diagnostic medical device. 50 FDA approval of AI‐based medical devices and algorithms since 2010 includes 64 devices and algorithms approved with a 510K clearance (85.9%), de novo clearance (12.5%) and premarket approval (1.6%), mostly in radiology, cardiology, and internal medicine. 51
Failures
Failures of clinical implementation are largely attributed to the lack of meaningful validations, 47 as well as legal, regulatory, and ethical issues, including patient privacy. Patients must be aware that their data may be used for algorithm development and/or stored in alternate locations. 48 , 52 These technologies must also address adaptation strategies to the healthcare landscape, including changes to the EHR, payments, diagnosis, and treatment. 48 Another important consideration is the potential “learning” bias that could impact many patients within diverse and under‐represented populations. 53 FDA approval of AI technologies is challenging due to the opacity of black‐box algorithms that (1) lack explanations for their predictions or recommendations and (2) permit continued learning of algorithms adapting to new information. 54 , 55
Opportunities
Opportunities include improving methods to validate AI technologies with implementation into healthcare workflows. Peer‐reviewed journals can assist in facilitating clinical validations by emphasizing important factors focusing on diagnostic accuracy and efficacy. 46 AI applications should be evaluated for usability by tracking keystrokes, mouse clicks, eye tracking, etc. 31 Multidisciplinary teams of researchers, providers, and regulators should be engaged to address regulatory hurdles of validation and continued algorithm learning while providing transparency. 54 Regulatory requirements should include algorithmic stewardship to provide ongoing oversight that ensures safety, effectiveness, and fairness in diverse populations. 53 In fact, clinical validations and successful implementation of AI into clinical workflows will enhance development of best practices. 47 AI has great potential to provide low‐cost, superior predictive models decreasing clinician cognitive load once issues of acceptability, fairness, and transparency have been addressed. 56
PUBLIC (POPULATION) HEALTH (T4)
Challenges
Challenges to the use of AI for population health include data quality and quantity, technology performance, as well as implementation and process change. Population health usually involves data from multiple resources at different levels of granularity. For example, to characterize and forecast an infectious disease outbreak in a region, we may get weekly, state‐level counts of laboratory confirmed cases from the National Notifiable Diseases Surveillance System 57 ; a daily number of cases from healthcare systems that may serve populations across state boundaries; population information in census tracts; internet search activities (zip code level); and interaction information from social media, like Facebook and Twitter (random samples from a region). Linking these data and handling the hierarchical relationships is still challenging.
Many other challenges arise in deploying AI for population health. For example, natural language processing (NLP), can automatically retrieve clinical and behavioral findings from free‐text medical reports, which greatly enriches the clinical features in epidemiology research (e.g., lifestyle exposure for Alzheimer’s disease 58 ). However, some misclassification is inevitable and may increase when applying NLP to new medical documents different from those that were used to train the tool. Furthermore, generalizability is still a challenge for machine learned models: a predictive model that is well‐developed using data from one region may not perform well in another region that has a different population served by a different healthcare system. 59 , 60 , 61 Regarding its application, the success of AI depends not just on science, but also on real‐world implementation and process change (e.g., public health decision making). 62
Successes
AI has been successfully applied to population surveillance. Examples include novel syndromic surveillance systems that use medical NLP and Bayesian network models to process free‐text medical reports, infer infectious respiratory disease outbreaks, and detect emerging diseases in populations. 63 , 64 The openly available public health intelligence system Health‐Map integrates data from disparate sources to produce a global view of infectious disease threats. 65 NLP tools can automatically analyze public tweets, monitor population medication abuse, 66 and adverse drug reactions. 67 Integrative spatiotemporal‐based analytical methods for population cohort studies of opioid poisoning in New York State reveal trends that reflect gender, age, economics, and location. 68
AI for epidemiology studies is also promising. Text knowledge engineering tools facilitate automatic cohort identification for research. 69 NLP tools successfully extract stage, histology, tumor grade, and therapies from the EHRs of patients with lung cancer 70 and unsupervised ML can facilitate discovery of latent disease clusters and patient subgroups from EHRs. 71 These discoveries may help researchers detect underlying patterns of diseases, which is a growing trend in epidemiology research.
Other successes involve environmental health, population genomics, death registration, etc. A recent study has determined with high accuracy the effects of severe aerial urban pollution on facial images. 72 ML‐based curation systems have classified genomic translational research through a Public Health Genomics Knowledge Base. 73 Finally, a deep neural network‐based model successfully coded causes of death in French death certificates. 74
Failures
No existing AI system is error‐free. For example, Google Flu Trends was wrong for 100 out of 108 weeks between 2011 and 2013, 75 and it missed the 2009 swine flu (H1N1) pandemic. System errors can lead to severe consequences for populations. Therefore, policy makers should still view AI technology as a complementary to human experts, not as replacements.
Opportunities
AI methods enable retrieval of health and non‐health data about populations and communities at different levels of granularity. For population‐level epidemiology studies, collaborative efforts demonstrate the value of AI, such as the Observational Health Data Sciences and Informatics Program (over half a billion patient records) 76 ; the Accrual to Clinical Trials Network (over 16 states) 77 ; the All of Us Program (over one million or more people from across the United States) 78 ; the PaTH Network 79 of the National Patient‐Centered Clinical Research Network (348 health systems and over 857K care providers); and the National Mesothelioma Virtual Bank includes over 10 years of specimens to support the nationwide research of this rare and lethal disease. 80 For healthcare policy and management, probabilistic record linkage of de‐identified administrative claims and electronic health records at the patient level 81 will facilitate cost‐effectiveness analysis. For population surveillance, social media, such as Twitter, is becoming a useful tool for population health surveillance and research involving the Affordable Care Act, health organizations, obesity, pet exposure, sexual health, transgender health, and vaccination. 82 For community intervention, AI technology is enabling smart interventions—using wearable devices, smart phones to monitor patients’ health status, 83 diet, 84 and medication adherence. 85 Moreover, ML may improve both predictive models and causal inference that can identify new and more effective approaches to reduce inequalities and improve population health. 86 Translational health disparities research is being enhanced by big data approaches, including linking structured data, harmonizing data elements, fostering citizen science, developing large longitudinal cohorts, and mining internet and social media data. 87
AI AND ML APPLICATIONS ACROSS THE CTSA CONSORTIUM
Not all AI/ML projects are reflected in the published literature. For example, operational projects within a health system may not reliably yield MEDLINE‐indexed publications. In addition, the field is moving so quickly that ongoing projects may be different from published work. To provide an additional perspective, we conducted a survey of 63 CTSA hubs about their AI/ML projects, including translational phase, funding, intended users, and AI approach (see Supplementary Material SB). The survey was administered using REDCap 88 hosted at The University of Texas Health Science Center at Houston. Informatics component directors at each CTSA site were the points of contact (POCs). POCs were contacted via email and asked to coordinate the survey response on behalf of their institution. After 2 weeks, POCs for hubs without responses were contacted again and asked to complete the survey.
As a complement to the survey, we retrieved all funded federal grants to the 63 CTSA hubs for the years 2011–2019 using NIH Research Portfolio Online Reporting Tools (RePORTER, https://reporter.nih.gov/, March 17, 2021). Years 2020–2021 were excluded because abstracts were not available via the RePORTER API. We then used the Medical Text Indexing software 89 , 90 to tag each result with MeSH terms that allowed us to categorize the projects according to the disease or syndrome addressed (if any) and the computational approach.
Results
Survey
Nineteen of 63 hubs responded to the survey (30% response rate) describing 63 distinct AI/ML projects. Multiple responses were allowed for each project. For example, a project could be funded by a combination of federal and institutional funds.
Responding hubs varied in the number of reported projects from one to 11 with a mean of four projects per hub. Federal funding was most common at 48.5% (Figure 2a). The most common translational phase was T2 (clinical research) at 40.2%, followed by T3 (clinical implementation) at 27.2% (Figure 2b). The most common AI approaches were supervised ML at 38.6% and deep learning at 34.2% (Figure 2c). The intended users were usually either clinicians at 44.6% or researchers at 32.3% (Figure 2d). The number of projects has been increasing over the past 8 years (Figure 2e).
FIGURE 2.
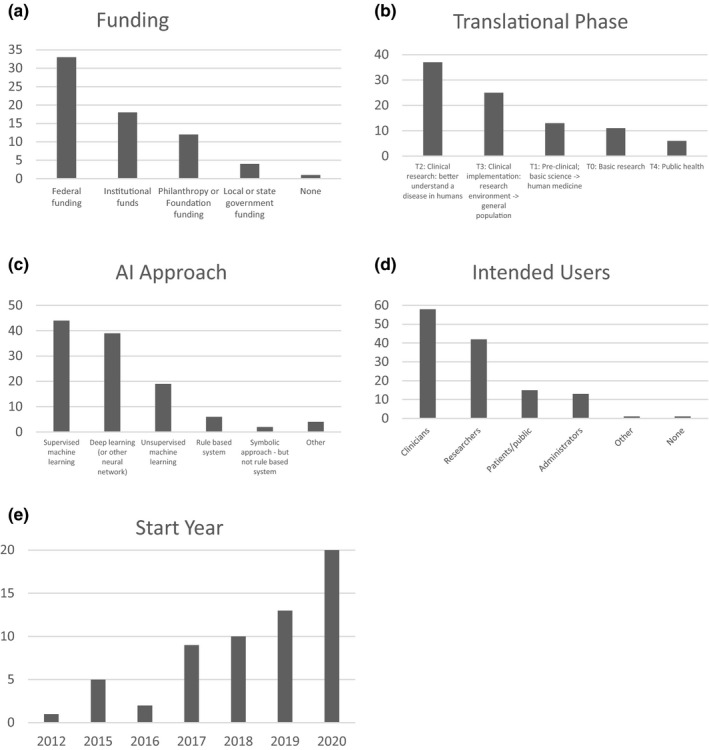
Artificial intelligence (AI) and machine learning (ML) project characteristics from Clinical and Translational Science Award (CTSA) institution survey (number of projects)
Some projects (24 of 63) could be mapped to disease categories (Figure 3). Infections and neoplasms were the most commonly addressed.
FIGURE 3.
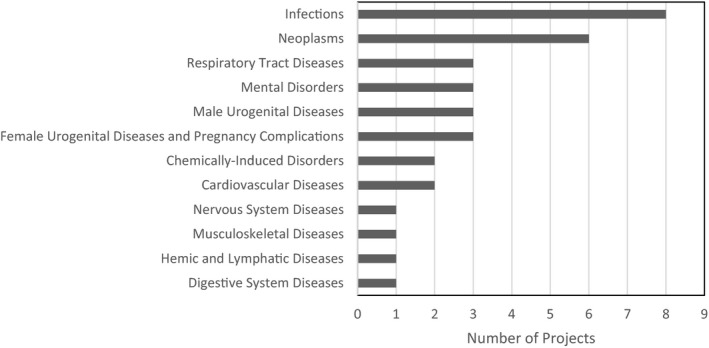
Disease categories of AI/ML Projects (Survey)
Federally funded projects (NIH RePORTER)
A total of 2604 funded projects were retrieved that contained either “machine learning” or “artificial intelligence” as keywords. Manual review of 200 results; 100 that were most likely to be related to AI and 100 that were least likely to be related showed that these keywords were accurate indicators of the project being related to AI.
Of these, 1379 could be mapped to disease categories. Again, a single project could address zero, one, or multiple disease categories. Nervous system diseases at 16.3% and mental disorders at 16.2% were most commonly addressed (Figure 4). Based on manual review, many projects that could not be mapped to a disease category were conducting basic science, such as “‐omics” work.
FIGURE 4.
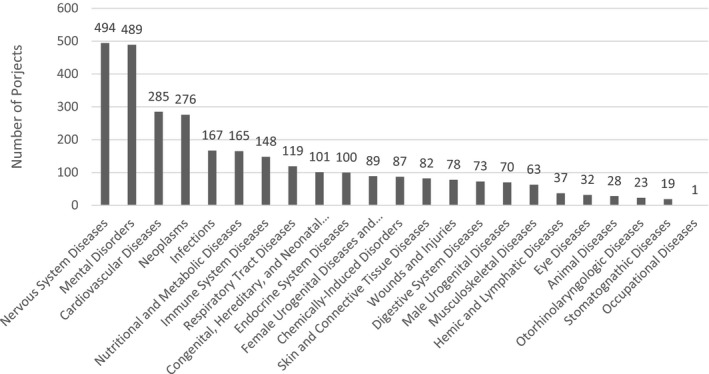
Disease categories of artificial intelligence/machine learning (AI/ML) Projects (Federally Funded Projects)
With respect to categories of computational approach, 697 of 2604 projects (26.8%) could be mapped to one or more computational category. The most common computational issues addressed were big data at 32.3% and deep learning at 30.0% (Figure 5).
FIGURE 5.
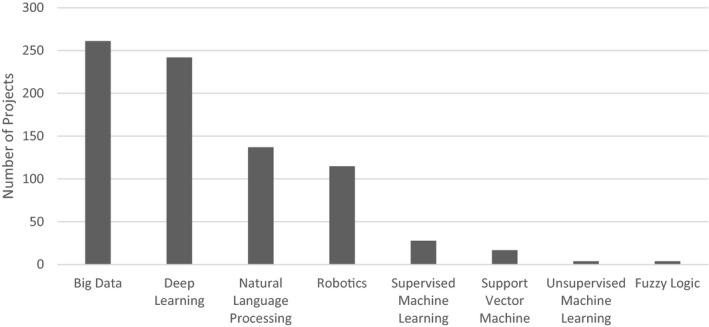
Computational domain of artificial intelligence/machine learning (AI/ML) Projects (Federally Funded Projects)
SUMMARY AND PATH FORWARD
In general, AI applications face the same challenges as other biomedical innovations, including validation using important outcomes (e.g., improved survival), rather than process measures or intermediate end points. Generalizability, or lack thereof, is a recurrent theme. Medical AI algorithms may work well under ideal conditions, but perform poorly in the real world, at another institution or with a different population. For example, surgical skin markings confused a deep learning algorithm for melanoma detection in which it classified benign nevi as malignant. 91 As another example, an AI system recommended “unsafe and incorrect” cancer treatments. 92 Similarly, a sepsis prediction algorithm implemented in a widely used EHR system performed poorly in practice. 34 Most algorithms were tested at a single institution, but validation studies at other institutions are rare.
The black box nature of algorithms, especially deep learning algorithms, makes it difficult to understand the reasoning of the AI algorithm. 93 With a weak explainable model, it is difficult to create actionable intervention plans. Moreover, weak explanation can complicate legal liability. For example, who is responsible when an algorithm recommends a treatment, but the patient does not respond well to that treatment? Alternatively, is a human clinician responsible for a poor outcome that might be attributed to not administering a treatment that is recommended by a “black box” AI system?
Medical AI algorithms are developed from clinical data and may faithfully learn all the biases of healthcare services and human clinicians. Using historical data to develop predictive models may encode racial and socioeconomic biases. 94 For example, an AI‐powered tool to prevent long hospital stays may discriminate against the most vulnerable patients, who need the most care. 95
For some applications, particularly those involving decision support, there are additional unanswered questions regarding acceptability. Although there are many technologies in routine use that are not fully understood (e.g., exact mechanism of action of some psychiatric drugs), it is not clear how best to make decision support tools based on statistical AI acceptable to clinicians. Questions such as: “Will the output have to include ‘explainable AI’?”; “Will it have to improve upon human judgment by a large margin?”; and “Does it have to meet certain generalizability criteria?” will have to be addressed. There are multiple systems that have shown parity with human judgment or even improvement upon human performance. 4 , 5 However, these are rarely (if ever) used in practice. 96 , 97
We recognize the distinction articulated between translational research (i.e., “endeavor to traverse a particular step of the translational process for a particular target or disease”) and translational science (i.e., “field of investigation focused on understanding the scientific and operational principles underlying each step of the translational process”). 98 We note that a specific project can contribute to both translational research and translational science. For example, the National COVID Cohort Collaborative (N3C, https://ncats.nih.gov/n3c), has already contributed to translational science (e.g., advancing regulatory structure for conducting large‐scale, data‐enabled research across institutions) and translational research (e.g., advancing our understanding of coronavirus disease [COVID]).
We also recognize that a great deal of relevant progress has been made outside of academia. Start‐up companies and the pharmaceutical industry have a great deal to contribute. However, these contributions can be challenging to evaluate and attribute based on the published literature as well as work done at academic medical centers. Further, there has been a great deal of work to apply AI/ML to identify candidate molecules for drug development, predict structure, and how proteins fold. For scope reasons, we excluded such “bioinformatics” work from this manuscript.
In conclusion, the clinical and translational research community may benefit from the strong message in Friedman’s “fundamental theorem” for biomedical informatics, “…a person working in partnership with an information (computing or AI) resource is “better” than that same person unassisted…” 99 Clearly, medicine is primed to take advantage of this “third wave” of AI and will assist the clinical and translational research to attaining the vision of a learning health ecosystem fueled by mobile computable knowledge. 100 Realizing the potential for AI in clinical and translational research will require collaboration between methodologists who design increasingly effective algorithms with clinicians and informaticians who can implement these algorithms into practice.
CONFLICT OF INTEREST
M.J.B. has a SFCOI in an AI in computational pathology company, SpIntellx, and is founder and stockholder. All other authors have declared no competing interests for this work.
Supporting information
Supplement S1
Supinfo S2
ACKNOWLEDGEMENTS
The authors thank Travis Holder at the Houston Academy of Medicine–Texas Medical Center library for help with the literature review as well as Dr. Jonathan Silverstein for helpful comments on earlier drafts of this paper.
Bernstam EV, Shireman PK, Meric‐Bernstam F, et al. Artificial intelligence in clinical and translational science: Successes, challenges and opportunities. Clin Transl Sci. 2022;15:309–321. doi: 10.1111/cts.13175
[Correction added on 12 November 2021, after first online publication: The surname of the author was corrected to Barapatre in this version.]
Funding information
This study was supported in part by National Center for Advancing Translational Sciences (NCATS) Synergy paper program through the Center for Leading Innovation and Collaboration (CLIC), NCATS grants UL1 TR000371 (Center for Clinical and Translational Sciences), UL1 TR001857, UL1 TR002645, NCATS and Office of the Director, NIH U01 TR002393, National Library of Medicine grants R01 LM011829 and K99 LM013383, National Institute of Aging P30 AG044271, PCORI CDRN‐1306‐04608, the Reynolds and Reynolds Professorship in Clinical Informatics, and the Cancer Prevention Research Institute of Texas (CPRIT) Data Science and Informatics Core for Cancer Research (RP170668).
REFERENCES
- 1. Pryor TA, Gardner RM, Clayton PD, Warner HR. The HELP System (pp. 19‐27). Proceedings of the Annual Symposium on Computer Applications in Medical Care; 1982. [Google Scholar]
- 2. Hripcsak G, Ludemann P, Pryor TA, Wigertz OB, Clayton PD. Rationale for the Arden Syntax. Comp Biomed Res Int J. 1994;27:291‐324. [DOI] [PubMed] [Google Scholar]
- 3. Shortliffe EH, Buchanan BG. Rule Based Expert Systems: The MYCIN Experiments of the Stanford Heuristic Programming Project. Addison‐Wesley; 1984. [Google Scholar]
- 4. Miller RA, Pople HE Jr, Myers JD. Internist‐1, an experimental computer‐based diagnostic consultant for general internal medicine. N Engl J Med. 1982;307:468‐476. [DOI] [PubMed] [Google Scholar]
- 5. de Dombal FT, Leaper DJ, Staniland JR, McCann AP, Horrocks JC. Computer‐aided diagnosis of acute abdominal pain. BMJ. 1972;2:9‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lenat DB, Prakash M, Shepherd M. CYC: using common sense knowledge to overcome brittleness and knowledge acquisition bottlenecks. AI Magazine. 1985;6:65‐85. [Google Scholar]
- 7. Esteva A, Kuprel B, Novoa RA, et al. Dermatologist‐level classification of skin cancer with deep neural networks. Nature. 2017;542:115‐118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Widrow B. An adaptive "ADALINE" neuron using chemical "MEMISTORS". 1960.
- 9. Maddox TM, Rumsfeld JS, Payne PRO. Questions for artificial intelligence in health care. JAMA. 2019;321:31‐32. [DOI] [PubMed] [Google Scholar]
- 10. Munn Z, Peters M, Stern C, Tufanaru C, Mcarthur A, Aromataris EC. Systematic review or scoping review? Guidance for authors when choosing between a systematic or scoping review approach. BMC Med Res Methodol. 2018;18:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Translational Science Spectrum: National Center for Advancing Translational Science; 2020. [cited November 13, 2020]. https://ncats.nih.gov/translation/spectrum.
- 12. Rifaioglu AS, Atas H, Martin MJ, Cetin‐Atalay R, Atalay V, Doğan T. Recent applications of deep learning and machine intelligence on in silico drug discovery: methods, tools and databases. Brief Bioinform. 2019;20:1878‐1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chang Y, Park H, Yang H‐J, et al. Cancer Drug Response Profile scan (CDRscan): a deep learning model that predicts drug effectiveness from cancer genomic signature. Sci Rep. 2018;8:8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chiu Y‐C, Chen H‐IH, Zhang T, et al. Predicting drug response of tumors from integrated genomic profiles by deep neural networks. BMC Med Genomics. 2019;12:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen J, Zhang L. A survey and systematic assessment of computational methods for drug response prediction. Brief Bioinform. 2021;22:232‐246. [DOI] [PubMed] [Google Scholar]
- 16. Zohora FT, Ziaur Rahman M, Tran NH, Xin L, Shan B, Li M. DeepIso: a deep learning model for peptide feature detection from LC‐MS map. Sci Rep. 2019;9:17168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. PIX Pharmaceuticals Announces Discontinuation of PRX‐00023 Clinical Development Program 2008. [cited November 13, 2020]. https://www.businesswire.com/news/home/20080320005247/en/EPIX‐Pharmaceuticals‐Announces‐Discontinuation‐PRX‐00023‐Clinical‐Development.
- 18. Tran NH, Qiao R, Xin L, Chen X, Shan B, Li M. Personalized deep learning of individual immunopeptidomes to identify neoantigens for cancer vaccines. Nat Mach Intell. 2020;2:764‐771. [Google Scholar]
- 19. Spagnolo DM, Al‐Kofahi Y, Zhu P, et al. Platform for quantitative evaluation of spatial intratumoral heterogeneity in multiplexed fluorescence images. Can Res. 2017;77:e71‐e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gerdes MJ, Sevinsky CJ, Sood A, et al. Highly multiplexed single‐cell analysis of formalin‐fixed, paraffin‐embedded cancer tissue. Proc Natl Acad Sci USA. 2013;110:11982‐11987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Uttam S, Stern AM, Sevinsky C, et al. Spatial domain analysis predicts risk of colorectal cancer recurrence and infers associated tumor microenvironment networks. Nat Commun. 2020;11:3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fassler DJ, Abousamra S, Gupta R, et al. Deep learning‐based image analysis methods for brightfield‐acquired multiplex immunohistochemistry images. Diagn Pathol. 2020;15:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sung NS, Crowley Jr WF, Genel M, et al. Central challenges facing the national clinical research enterprise. JAMA. 2003;289:1278‐1287. [DOI] [PubMed] [Google Scholar]
- 24. Embi PJ, Payne PR. Clinical research informatics: challenges, opportunities and definition for an emerging domain. J Am Med Inform Assoc. 2009;16:316‐327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Getz KA, Campo RA. New benchmarks characterizing growth in protocol design complexity. Ther Innov Regul Sci. 2018;52:22‐28. [DOI] [PubMed] [Google Scholar]
- 26. Eisenstein EL, Lemons PW, Tardiff BE, Schulman KA, Jolly MK, Califf RM. Reducing the costs of phase III cardiovascular clinical trials. Am Heart J. 2005;149:482‐488. [DOI] [PubMed] [Google Scholar]
- 27. Malakoff D. Clinical trials and tribulations. Spiraling Costs Threaten Gridlock. Science. 2008;322:210‐213. [DOI] [PubMed] [Google Scholar]
- 28. Haynes B. Can it work? Does it work? Is it worth it? The testing of healthcare interventions is evolving. BMJ. 1999;319:652‐653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. FDA . Framework for FDA’s Real‐World Evidence Program. 2018.
- 30. Waring J, Lindvall C, Umeton R. Automated machine learning: Review of the state‐of‐the‐art and opportunities for healthcare. Artif Intell Med. 2020;104:101822. [DOI] [PubMed] [Google Scholar]
- 31. Rundo L, Pirrone R, Vitabile S, Sala E, Gambino O. Recent advances of HCI in decision‐making tasks for optimized clinical workflows and precision medicine. J Biomed Inform. 2020;108:103479. [DOI] [PubMed] [Google Scholar]
- 32. Fumeaux NF, Ebrahim S, Coughlin BF, et al. Accurate detection of spontaneous seizures using a generalized linear model with external validation. Epilepsia. 2020;61:1906‐1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Caccamisi A, Jorgensen L, Dalianis H, Rosenlund M. Natural language processing and machine learning to enable automatic extraction and classification of patients’ smoking status from electronic medical records. Upsala J Med Sci. 2020;125:316‐324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wong A, Otles E, Donnelly JP, et al. External validation of a widely implemented proprietary sepsis prediction model in hospitalized patients. JAMA Intern Med. 2021;181:1065‐1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu P, Fu BO, Yang SX, Deng L, Zhong X, Zheng H. Optimizing survival analysis of XGBoost for ties to predict disease progression of breast cancer. IEEE Transac Biomed Eng. 2021;68:148‐160. [DOI] [PubMed] [Google Scholar]
- 36. FDA . Title 21–Food and Drugs, Chapter I Food and Drug Administration Department of Health and Human Services. Part 11 Electronic Records; Electronic Signatures. 1997.
- 37. FDA . Patient‐reported outcome measures: use in medical product development to support labeling claims guidance for industry. 2009. [DOI] [PMC free article] [PubMed]
- 38. Aboulelenein S, Williams T, Baldner J, Zozus MN. Analysis of professional competencies for the clinical research data management profession. Data Basics. 2020;26:6‐17. [DOI] [PubMed] [Google Scholar]
- 39. Wilkinson M, Young R, Harper B, Machion B, Getz K. Baseline assessment of the evolving 2017 eClinical landscape. Ther Innov Regul Sci. 2019;53:71‐80. [DOI] [PubMed] [Google Scholar]
- 40. Eisenstein E, Zozus MN, Sanns W. Beyond EDC. J Soc Clin Data Manag. 2021;1(1):1–22. [Google Scholar]
- 41. Fouarge E, Monseur A, Boulanger B, et al. Hierarchical Bayesian modelling of disease progression to inform clinical trial design in centronuclear myopathy. Orphanet J Rare Dis. 2021;16:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Richard E, Reddy B. Text classification for clinical trial operations: evaluation and comparison of natural language processing techniques. Ther Innov Regul Sci. 2021;55:447‐453. [DOI] [PubMed] [Google Scholar]
- 43. Godfrey A, Vandendriessche B, Bakker JP, et al. Fit‐for‐purpose biometric monitoring technologies: leveraging the laboratory biomarker experience. Clin Transl Sci. 2021;14:62‐74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tabuchi H, Masumoto H. Objective evaluation of allergic conjunctival disease (with a focus on the application of artificial intelligence technology). Allergol Int. 2020;69:505‐509. [DOI] [PubMed] [Google Scholar]
- 45. Zeng K, Pan Z, Xu Y, Qu Y. An ensemble learning strategy for eligibility criteria text classification for clinical trial recruitment: algorithm development and validation. JMIR Med Inform. 2020;8:e17832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Park SH, Kressel HY. Connecting technological innovation in artificial intelligence to real‐world medical practice through rigorous clinical validation: what peer‐reviewed medical journals could do. J Korean Med Sci. 2018;33:e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sendak MP, Ratliff W, Sarro D, et al. Real‐world integration of a sepsis deep learning technology into routine clinical care: implementation study. JMIR Med Inform. 2020;8:e15182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Noorbakhsh‐Sabet N, Zand R, Zhang Y, Abedi V. Artificial intelligence transforms the future of health care. Am J Med. 2019;132:795‐801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wiens J, Saria S, Sendak M, et al. Do no harm: a roadmap for responsible machine learning for health care. Nat Med. 2019;25:1337‐1340. [DOI] [PubMed] [Google Scholar]
- 50. Abramoff MD, Lavin PT, Birch M, Shah N, Folk JC. Pivotal trial of an autonomous AI‐based diagnostic system for detection of diabetic retinopathy in primary care offices. NPJ Digit Med. 2018;1:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Benjamens S, Dhunnoo P, Mesko B. The state of artificial intelligence‐based FDA‐approved medical devices and algorithms: an online database. NPJ Digit Med. 2020;3:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mitchell C, Ploem C. Legal challenges for the implementation of advanced clinical digital decision support systems in Europe. J Clin Transl Res. 2018;3:424‐430. [PMC free article] [PubMed] [Google Scholar]
- 53. Eaneff S, Obermeyer Z, Butte AJ. The case for algorithmic stewardship for artificial intelligence and machine learning technologies. JAMA. 2020;324(14):1397. [DOI] [PubMed] [Google Scholar]
- 54. Price WN. Big data and black‐box medical algorithms. Sci Transl Med. 2018;10:eaao5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. London AJ. Artificial intelligence and black‐box medical decisions: accuracy versus explainability. Hastings Cent Rep. 2019;49:15‐21. [DOI] [PubMed] [Google Scholar]
- 56. Yu KH, Beam AL, Kohane IS. Artificial intelligence in healthcare. Nat Biomed Eng. 2018;2:719‐731. [DOI] [PubMed] [Google Scholar]
- 57. CDC, A . National Notifiable Diseases Surveillance System (NNDSS).
- 58. Zhou X, Wang Y, Sohn S, Therneau TM, Liu H, Knopman DS. Automatic extraction and assessment of lifestyle exposures for Alzheimer’s disease using natural language processing. Int J Med Informatics. 2019;130:103943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Carroll RJ, Thompson WK, Eyler AE, et al. Portability of an algorithm to identify rheumatoid arthritis in electronic health records. J Am Med Inform Assoc. 2012;19:e162‐e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ye Y, Wagner MM, Cooper GF, et al. A study of the transferability of influenza case detection systems between two large healthcare systems. PLoS One. 2017;12:e0174970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Couzin‐Frankel J. Medicine contends with how to use artificial intelligence. Science. 2019;364(6446):1119–1120. [DOI] [PubMed] [Google Scholar]
- 62. Talby D. Three insights from Google's ‘Failed’ Field Test To Use AI For Medical Diagnosis. https://www.forbes.com/sites/forbestechcouncil/2020/06/09/three‐insights‐from‐googles‐failed‐field‐test‐to‐use‐ai‐for‐medical‐diagnosis/?sh=4a897fddbac4. 2020; September 26, 2020.
- 63. Aronis JM, Ferraro JP, Gesteland PH, et al. A Bayesian approach for detecting a disease that is not being modeled. PLoS One. 2020;15:e0229658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cooper GF, Villamarin R, (Rich) Tsui F‐C, Millett N, Espino JU, Wagner MM. A method for detecting and characterizing outbreaks of infectious disease from clinical reports. J Biomed Inform. 2015;53:15‐26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Brownstein JS, Freifeld CC, Madoff LC. Digital disease detection—harnessing the Web for public health surveillance. N Engl J Med. 2009;360:2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sarker A, O’Connor K, Ginn R, et al. Social media mining for toxicovigilance: automatic monitoring of prescription medication abuse from Twitter. Drug Saf. 2016;39:231‐240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Korkontzelos I, Nikfarjam A, Shardlow M, Sarker A, Ananiadou S, Gonzalez GH. Analysis of the effect of sentiment analysis on extracting adverse drug reactions from tweets and forum posts. J Biomed Inform. 2016;62:148‐158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Schoenfeld ER, Leibowitz GS, Wang YU, et al. Geographic, temporal, and sociodemographic differences in opioid poisoning. Am J Prev Med. 2019;57:153‐164. [DOI] [PubMed] [Google Scholar]
- 69. Weng C, Wu X, Luo Z, et al. EliXR: an approach to eligibility criteria extraction and representation. J Am Med Inform Assoc. 2011;18:i116‐i124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang L, Luo L, Wang Y, Wampfler J, Yang P, Liu H. Natural language processing for populating lung cancer clinical research data. BMC Med Inform Decis Mak. 2019;19:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wang Y, Yiqing Z, Therneau T, et al. Unsupervised machine learning for the discovery of latent disease clusters and patient subgroups using electronic health records. J Biomed Inform. 2020;102:103364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhang Y, Jiang R, Kezele I, et al. A new procedure, free from human assessment, that automatically grades some facial skin signs in men from selfie pictures. Application to changes induced by a severe aerial chronic urban pollution. Int J Cosmet Sci. 2020;42:185‐197. [DOI] [PubMed] [Google Scholar]
- 73. Hsu Y‐Y, Clyne M, Wei C‐H, Khoury MJ, Lu Z. Using deep learning to identify translational research in genomic medicine beyond bench to bedside. Database. 2019;2019:baz010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Falissard L, Morgand C, Roussel S, et al. A deep artificial neural network− based model for prediction of underlying cause of death from death certificates: algorithm development and validation. JMIR Med Inform. 2020;8:e17125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lazer D, Kennedy R, King G, Vespignani A. The parable of Google Flu: traps in big data analysis. Science. 2014;343:1203‐1205. [DOI] [PubMed] [Google Scholar]
- 76. Hripcsak G, Duke JD, Shah NH, et al. Observational Health Data Sciences and Informatics (OHDSI): opportunities for observational researchers. Stud Health Technol Inform. 2015;216:574. [PMC free article] [PubMed] [Google Scholar]
- 77. Visweswaran S, Becich MJ, D’Itri VS, et al. Accrual to clinical trials (ACT): a clinical and translational science award consortium network. JAMIA Open. 2018;1:147‐152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Investigators . The “All of Us” research program. N Engl J Med. 2019;381:668‐676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Amin W, Tsui F, Borromeo C, et al. PaTH: towards a learning health system in the Mid‐Atlantic region. J Am Med Inform Assoc. 2014;21:633‐636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Amin W, Parwani AV, Schmandt L, et al. National Mesothelioma Virtual Bank: a standard based biospecimen and clinical data resource to enhance translational research. BMC Cancer. 2008;8:1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hejblum BP, Weber GM, Liao KP, et al. Probabilistic record linkage of de‐identified research datasets with discrepancies using diagnosis codes. Sci Data. 2019;6:180298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Sinnenberg L, Buttenheim AM, Padrez K, Mancheno C, Ungar L, Merchant RM. Twitter as a tool for health research: a systematic review. Am J Public Health. 2017;107:e1‐e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Li J, Ma Q, Chan AH, Man S. Health monitoring through wearable technologies for older adults: smart wearables acceptance model. Appl Ergon. 2019;75:162‐169. [DOI] [PubMed] [Google Scholar]
- 84. Ipjian ML, Johnston CS. Smartphone technology facilitates dietary change in healthy adults. Nutrition. 2017;33:343‐347. [DOI] [PubMed] [Google Scholar]
- 85. Khan A, Khusro S. Smart assist: Smartphone‐based drug compliance for elderly people and people with special needs. In: Fazlullah K, Mian Ahmad J, Muhammad A, eds. Applications of Intelligent Technologies in Healthcare (pp. 99‐108). Springer; 2019. [Google Scholar]
- 86. Glymour MM, Osypuk TL, Rehkopf DH. Invited commentary: off‐roading with social epidemiology—exploration, causation, translation. Am J Epidemiol. 2013;178:858‐863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Breen N, Berrigan D, Jackson JS, et al. Translational health disparities research in a data‐rich world. Health Equity. 2019;3:588‐600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)–a metadata‐driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377‐381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Mork J, Aronson A, Demner‐Fushman D. 12 years on ‐ Is the NLM medical text indexer still useful and relevant? J Biomed Semant. 2017;8:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Rae AR, Pritchard DO, Mork JG, Demner‐Fushman D. Automatic MeSH indexing: revisiting the subheading attachment problem. AMIA … Annual Symposium proceedings. AMIA Symposium. 2020:1031‐1040. [PMC free article] [PubMed]
- 91. Winkler JK, Fink C, Toberer F, et al. Association between surgical skin markings in dermoscopic images and diagnostic performance of a deep learning convolutional neural network for melanoma recognition. JAMA Dermatol. 2019;155:1135‐1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Casey Ross IS. IBM’s Watson supercomputer recommended ‘unsafe and incorrect’ cancer treatments, internal documents show. https://www.statnews.com/wp‐content/uploads/2018/09/IBMs‐Watson‐recommended‐unsafe‐and‐incorrect‐cancer‐treatments‐STAT.pdf. 2018; September 26, 2020.
- 93. Caruana R. Friends don't let friends deploy black‐box models: the importance of intelligibility in machine learning. Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining. 2019: 3174.
- 94. Obermeyer Z, Powers B, Vogeli C, Mullainathan S. Dissecting racial bias in an algorithm used to manage the health of populations. Science. 2019;366:447‐453. [DOI] [PubMed] [Google Scholar]
- 95. Blasimme A, Vayena E. The Ethics of AI in Biomedical research, patient care and public health. Patient Care and Public Health (April 9, 2019). Oxford Handbook of Ethics of Artificial Intelligence, Forthcoming. (2019).
- 96. Kelly CJ, Karthikesalingam A, Suleyman M, Corrado G, King D. Key challenges for delivering clinical impact with artificial intelligence. BMC Med. 2019;17:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. He J, Baxter SL, Xu J, Xu J, Zhou X, Zhang K. The practical implementation of artificial intelligence technologies in medicine. Nat Med. 2019;25:30‐36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. PAR‐21‐293 Clinical and Translational Science Award (UM1): National Institutes of Health; 2021. https://grants.nih.gov/grants/guide/pa‐files/PAR‐21‐293.html.
- 99. Friedman CP. A "fundamental theorem" of biomedical informatics. J Am Med Inform Assoc. 2009;16:169‐170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Adler‐Milstein J, Nong P, Friedman CP. Preparing healthcare delivery organizations for managing computable knowledge. Learn Health Syst. 2019;3:e10070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement S1
Supinfo S2


