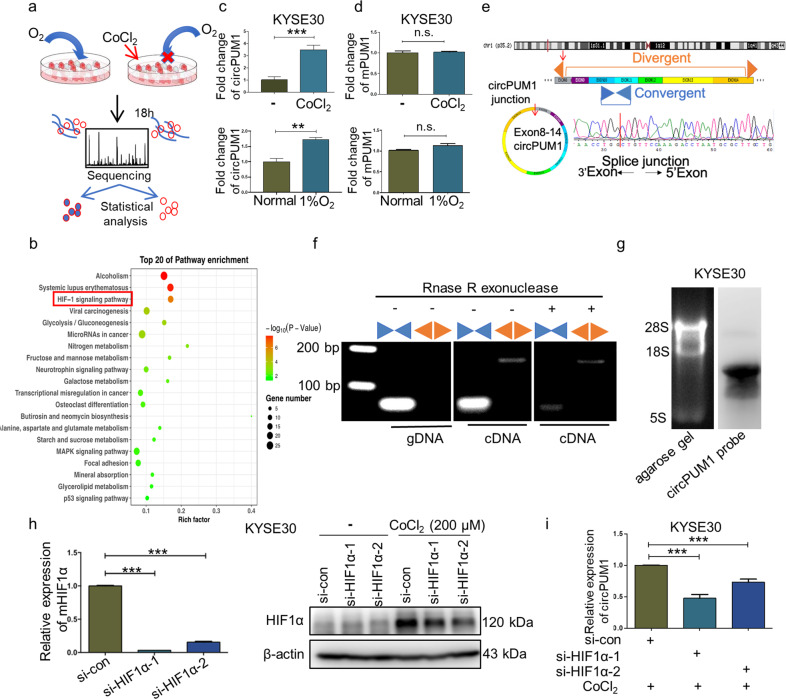
Fig. 1.
High-throughput identification of circPUM1 in CoCl2-induced intracellular hypoxic-like microenvironment model. a Intracellular hypoxic-like microenvironment model creation and sequencing strategy. b Pathway enrichment analysis of differentially expressed genes. c RT-qPCR assay to determine circPUM1 expression level after cells were treated with 200 μM CoCl2 for18 h or incubated in 1% oxygen for 24 h. d RT-qPCR assay to determine PUM1 mRNA expression level after cells were treated with 200 μM CoCl2 for 18 h or incubated in 1% oxygen for 24 h. e Sequencing analysis of head-to-tail splicing junction in circPUM1. f Divergent and convergent primers were used to amplify circPUM1 and mPUM1 from gDNA, cDNA without RNase R treatment before RT-PCR and cDNA with RNase R treatment before RT-PCR in KYSE30 cell, respectively. g Northern blotting analysis of circPUM1 in KYSE30 cell. h RT-qPCR and western blot analyses for the knockdown efficiency of si-HIF1α in KYSE30 cell. i RT-qPCR assay for circPUM1 expression following the depletion of HIF1α and the treatment with 200 μM CoCl2 for 18 h in KYSE30 cell