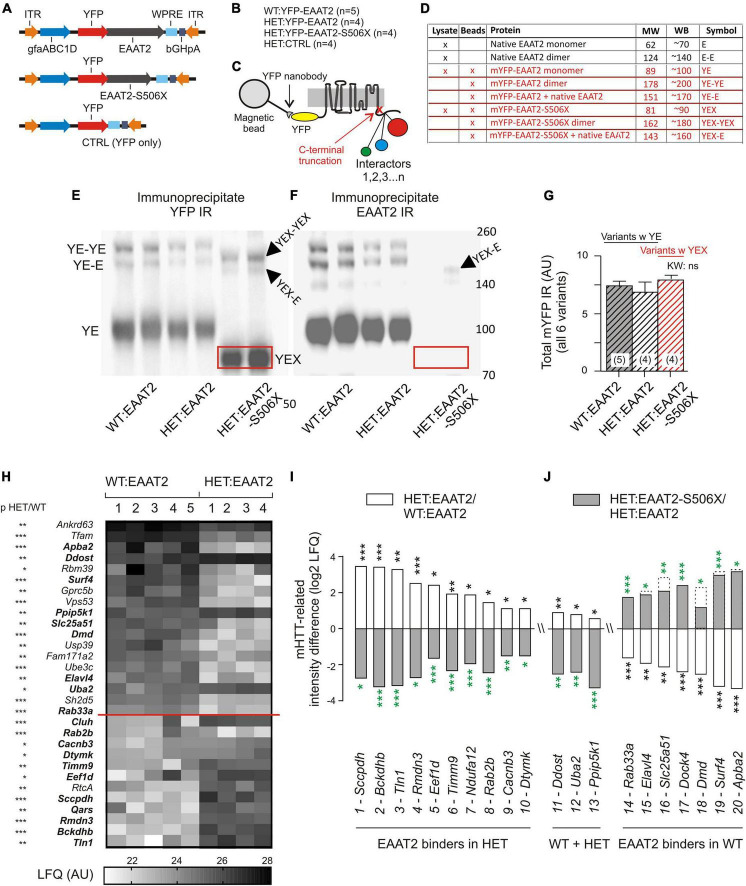
FIGURE 2.
mHTT- and S506X-related changes in the abundance of YFP-EAAT2 binders. (A) Injected vectors. (B) Animal groups. (C) Scheme of immunoprecipitation experiment. (D) List of EAAT2 bands to be detected in the Western blots (WB) from the beads preparation (this figure) or lysates (Figure 3). (E,F) Western blot samples prepared from the beads YFP-immunoprecipitate. Note dimer bands and shift of the EAAT2-S506X band (YEX, boxed). (G) Similar amounts of mYFP immunoreactivity (YE or YEX bands) pulled down in the three animal test groups. (H) Heat-map plot illustrating the mHTT-related differences between WT:EAAT2 and HET:EAAT2. The signal from each animal was normalized to the mYFP median intensity value in a histogram constructed from all samples. The inter-group intensities were compared by a two-tailed t-test. The asterisks next to the listed genes denote the significance level of the difference. The proteins are listed with their gene names and sorted according to their abundance in WT:EAAT2. (I,J) Plot illustrating the recovery potential of significant EAAT2 binders. The LFQ intensity difference denotes the log2 difference between the compared groups. Compared were WT:EAAT2 vs. HET:EAAT2 (empty bars with black asterisks) and HET:EAAT2-S506X vs. HET:EAAT2 (filled bars with green asterisks). The dashed bars indicate the corresponding WT levels. An up-regulation is shown as upward bar, a down-regulation as downward bar. *p < 0.05, **p < 0.01, ***p < 0.001.