Figure 2.
Hybrid origins of the cutaneous Himachal isolate
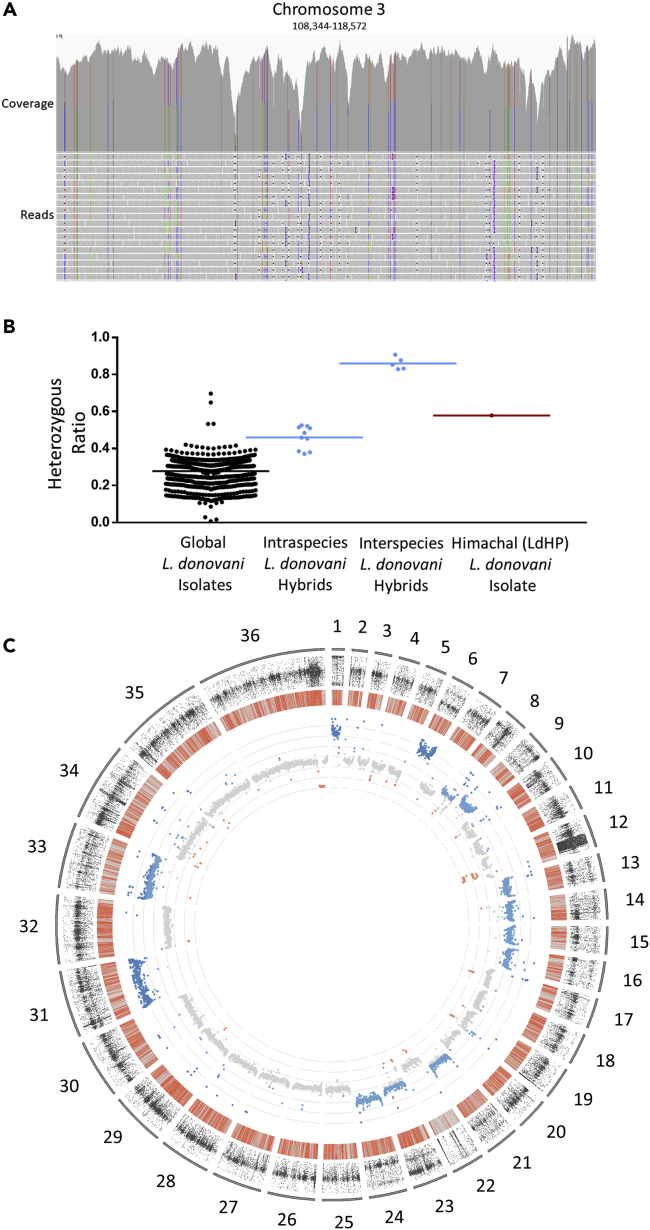
(A) Representative alignment of a 10 kb window on chromosome 3 (x axis) for the cutaneous isolate (LdHP) showing the density of heterozygous polymorphisms. The individual reads (y axis) are aligned in the bottom panel. Perfect base matches are shown in gray, mismatches are colored according to the corresponding polymorphic base (ATCG). The upper panel shows the resulting coverage from all alignments on this region of the genome with the same color coding.
(B) The heterozygous polymorphism ratio of the Himachal cutaneous isolate is consistent with an intraspecies hybrid genotype. The frequency of heterozygous mutations in the cutaneous isolate was compared to those found in the worldwide population of L. donovani, and two baseline hybrid population frequencies: an intraspecies hybrid focus from distinct Ethiopian clades and an interspecies hybrid parasite focus in Sri Lanka previously characterized.
(C) Whole genome sequencing coverage depth to determine chromosomal copy number. Mean coverage of each gene is plotted along the chromosomes and colored according to median copy number (gray), decrease (red) or an increase in copy number (blues). The average of all chromosomes serves as the baseline for diploid levels, normally tetraploid chromosome 31 serves as the baseline for tetraploidy. Chromosomes one and six show increased copy number of all genes along their length consistent with a chromosomal amplification resulting in tetraploidy. Chromosomes 8,9,13–16,21,23,26 & 33 show an intermediate increase in copy numbers across the entire chromosomes consistent with an amplification resulting in triploidy (inner track). The location of all polymorphisms identified compared to the reference LdCL genome are shown in the middle track. Homozygous polymorphisms are colored in gray, heterozygous (likely hybrid) polymorphisms are colored in red. Heterozygous polymorphisms are spread evenly across the entire genome with the exception of chromosome 22 which appears mostly homozygous. The frequency of heterozygous SNP is plotted at every heterozygous loci (outer track) and shows polyploid chromosomes moving from an even ∼50/50 allele distribution to a bimodal (~33/66 or ∼25/75) distribution along the chromosomes with increased ploidy.