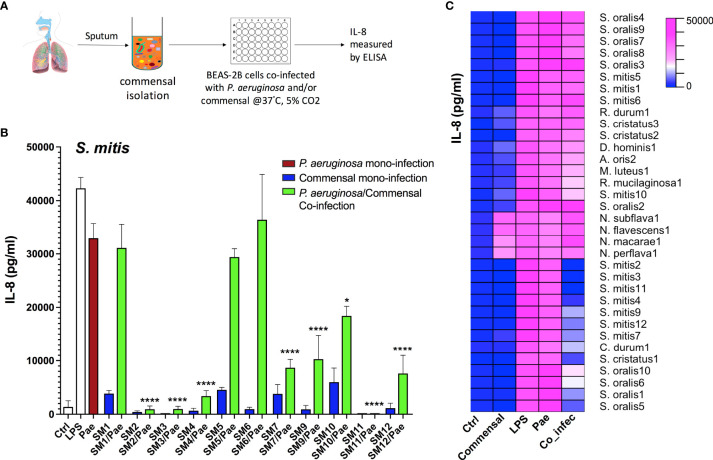
Figure 1.
Multiple Streptococcus strains reduce P. aeruginosa triggered inflammatory responses as indicated by the reduction of IL-8 in co-infection compared to that of P. aeruginosa mono-infection in BEAS-2B cells. (A) Cell culture based experimental workflow of the screening study. (B&C) BEAS-2B cells were non-infected (Ctrl), or mono-infected with P. aeruginosa PAO1 (labelled as Pae) or one CF-microbiome isolated commensal strain, or co-infected with both of them. LPS stimulation is used as positive control. Release of IL-8 into the supernatant was assessed after 2 h of infection followed by overnight incubation. (B) Representative results from Streptococcus mitis strains (“SM+number” is the name of self-isolated strains). Data are presented as Mean ± SEM (N=6 experiments). *p < 0.05, ****p < 0.0001 indicate significant difference between co-infection and P. aeruginosa mono-infection. (C) The major results are summarized as a heatmap. Ctrl, non-infected control samples; Commensal, commensal mono-infection; LPS, LPS stimulation as positive control; Pae, P. aeruginosa PAO1 mono-infection; Co_infec, P. aeruginosa/commensal co-infection. S., Streptococcus; N., Neisseria; D., Dermabacter; A., Actinomyces; M., Micrococcus; R., Rothia; C., Corynebacterium. Strain name such as SM4 is labelled as S. mitis4; N=6 experiments.